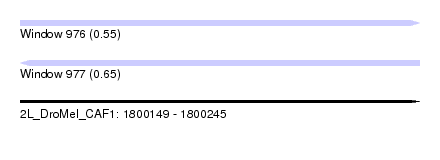
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,800,149 – 1,800,245 |
| Length | 96 |
| Max. P | 0.649181 |

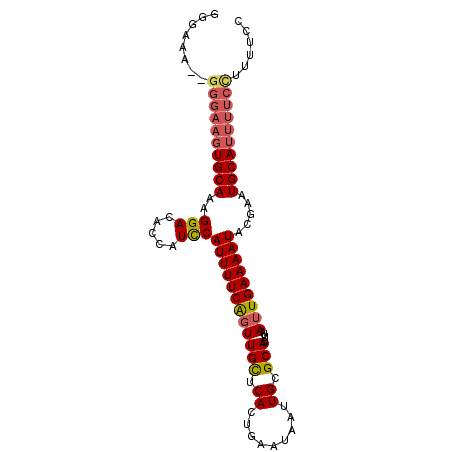
| Location | 1,800,149 – 1,800,245 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 80.84 |
| Mean single sequence MFE | -21.24 |
| Consensus MFE | -15.61 |
| Energy contribution | -16.83 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1800149 96 + 22407834 GGAAAAGGAAAAUGCAUUCGUAUUUUCAAUUAAAGUGCGCAAUUAUUCAGUGAGCAACCGAAAAUGGAUGGUGUCCUUUGCACUUCCCACUUUCCC ((.....((((((((....)))))))).......((.(((.........))).))..))((((.(((..(((((.....)))))..))).)))).. ( -22.90) >DroSim_CAF1 2288 88 + 1 GGAAAAGGAAAAUGCAUUCGUAUUUUCAAUUAAAGUGCGCAAUUAUUCAGUGAGCAACUGAAAAUGGAUGGUGUCCUUUGCACUUC--------CC ((((...((((((((....)))))))).......((((.......((((((.....))))))...((((...))))...)))))))--------). ( -24.90) >DroEre_CAF1 9948 88 + 1 GGUCGAAGAAAGUGCAUUUGUAUUUUCAAUUAAAGUGCGCACUUAUUCAGUGAACAAAUGAAAAUGAA----GUCCUUUGCAU---CC-AUUUCCC ((.(((((...((((((((.............))))))))((((.((((.........))))....))----)).)))))...---))-....... ( -15.92) >consensus GGAAAAGGAAAAUGCAUUCGUAUUUUCAAUUAAAGUGCGCAAUUAUUCAGUGAGCAACUGAAAAUGGAUGGUGUCCUUUGCACUUCCC__UUUCCC .......((((((((....)))))))).....((((((.......((((((.....))))))...(((.....)))...))))))........... (-15.61 = -16.83 + 1.23)

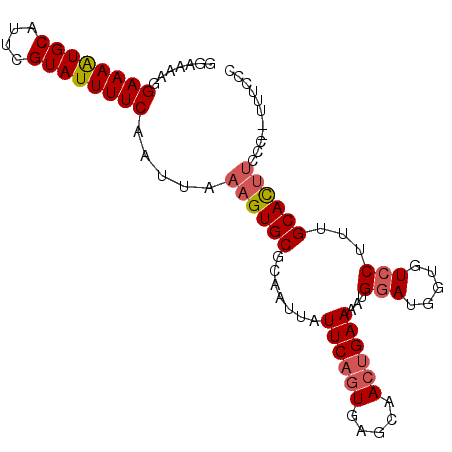
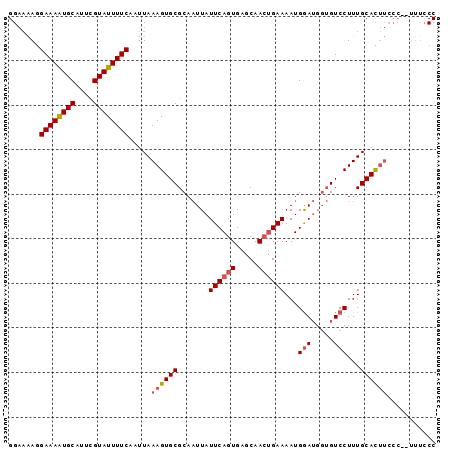
| Location | 1,800,149 – 1,800,245 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 80.84 |
| Mean single sequence MFE | -21.57 |
| Consensus MFE | -14.37 |
| Energy contribution | -15.93 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1800149 96 - 22407834 GGGAAAGUGGGAAGUGCAAAGGACACCAUCCAUUUUCGGUUGCUCACUGAAUAAUUGCGCACUUUAAUUGAAAAUACGAAUGCAUUUUCCUUUUCC .((((((.((((((((((..(((.....)))((((((((((((.((.(.....).)).))).....))))))))).....)))))))))))))))) ( -27.80) >DroSim_CAF1 2288 88 - 1 GG--------GAAGUGCAAAGGACACCAUCCAUUUUCAGUUGCUCACUGAAUAAUUGCGCACUUUAAUUGAAAAUACGAAUGCAUUUUCCUUUUCC ((--------((((((((..(((.....)))((((((((((((.((.(.....).)).))).....))))))))).....))))))))))...... ( -23.40) >DroEre_CAF1 9948 88 - 1 GGGAAAU-GG---AUGCAAAGGAC----UUCAUUUUCAUUUGUUCACUGAAUAAGUGCGCACUUUAAUUGAAAAUACAAAUGCACUUUCUUCGACC .((((((-((---(..(....)..----)))))))))..........((((.(((((((.....................)))))))..))))... ( -13.50) >consensus GGGAAA__GGGAAGUGCAAAGGACACCAUCCAUUUUCAGUUGCUCACUGAAUAAUUGCGCACUUUAAUUGAAAAUACGAAUGCAUUUUCCUUUUCC ........((((((((((..(((.....)))((((((((((((.((.........)).))).....))))))))).....))))))))))...... (-14.37 = -15.93 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:31 2006