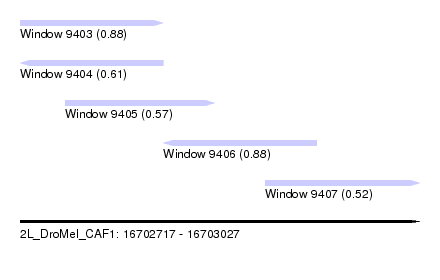
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,702,717 – 16,703,027 |
| Length | 310 |
| Max. P | 0.884830 |

| Location | 16,702,717 – 16,702,828 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.81 |
| Mean single sequence MFE | -23.94 |
| Consensus MFE | -19.99 |
| Energy contribution | -20.30 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16702717 111 + 22407834 CGAAAAAAGUGCAGUCCGGUCGUAAAACAUU-----UCCA----UAUAUCUGGCGGACAGUAAAUAAUAAAAAAAAAAAGCUACGACAGCAGCUUGUCGAGUCCACGCGACACAUGAAAA ..................(((((........-----.(((----......))).(((((((..................))).((((((....)))))).))))..)))))......... ( -19.47) >DroSec_CAF1 157406 102 + 1 CGAAAAAAGUGCAGUCCGGUCGUAAAACAUU-----UCCA----UAUAUCUGGCGGACAAUA---------AAAAAAAAGCUACGACAGCAGCUUGUCGCGUCCACGCGACACAUAAAAA ..........((.(((((((......))...-----.(((----......))))))))....---------........(((.....))).)).(((((((....)))))))........ ( -24.00) >DroSim_CAF1 157336 106 + 1 CGAAAAAAGUGCAGUCCGGUCGUAAAACAUU-----UCCAGAUGUAUAUCUGGCGGAUAAUA---------AAAAAAAAGCUACGACAGCAGCUUGUCGCGUCCACGCGACACAUAAAAA ..........((.(((((((......))...-----.((((((....)))))))))))....---------........(((.....))).)).(((((((....)))))))........ ( -27.10) >DroEre_CAF1 158799 109 + 1 CGAAAAAAGUGCAGUCCGGUCGUAAAACAUUUCAUUUCCA----UAUAUCUGGCGGACAAUA-------AUAAAAAAAAGCAGCGACAGCAGCUUGUCGCGUCCACGCGACACAUAAAAA .............(((((((......)).........(((----......))))))))....-------........((((.((....)).))))((((((....))))))......... ( -25.20) >consensus CGAAAAAAGUGCAGUCCGGUCGUAAAACAUU_____UCCA____UAUAUCUGGCGGACAAUA_________AAAAAAAAGCUACGACAGCAGCUUGUCGCGUCCACGCGACACAUAAAAA ..........((.(((((((......)).........(((..........)))))))).....................(((.....))).)).(((((((....)))))))........ (-19.99 = -20.30 + 0.31)

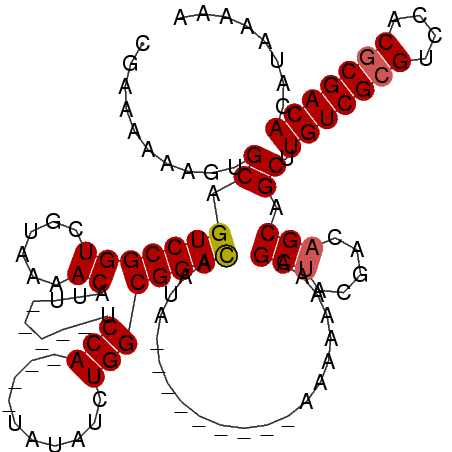
| Location | 16,702,717 – 16,702,828 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.81 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -22.16 |
| Energy contribution | -22.73 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16702717 111 - 22407834 UUUUCAUGUGUCGCGUGGACUCGACAAGCUGCUGUCGUAGCUUUUUUUUUUUAUUAUUUACUGUCCGCCAGAUAUA----UGGA-----AAUGUUUUACGACCGGACUGCACUUUUUUCG ((((((((((((..((((((.....(((((((....)))))))...................))))))..))))))----))))-----))(((....(....)....)))......... ( -26.85) >DroSec_CAF1 157406 102 - 1 UUUUUAUGUGUCGCGUGGACGCGACAAGCUGCUGUCGUAGCUUUUUUUU---------UAUUGUCCGCCAGAUAUA----UGGA-----AAUGUUUUACGACCGGACUGCACUUUUUUCG ......(((((((((....))))))(((((((....)))))))......---------....((((((((......----))).-----...(((....)))))))).)))......... ( -27.60) >DroSim_CAF1 157336 106 - 1 UUUUUAUGUGUCGCGUGGACGCGACAAGCUGCUGUCGUAGCUUUUUUUU---------UAUUAUCCGCCAGAUAUACAUCUGGA-----AAUGUUUUACGACCGGACUGCACUUUUUUCG ......(((((((((....))))))(((((((....)))))))......---------.....((((((((((....)))))).-----...(((....)))))))..)))......... ( -29.10) >DroEre_CAF1 158799 109 - 1 UUUUUAUGUGUCGCGUGGACGCGACAAGCUGCUGUCGCUGCUUUUUUUUAU-------UAUUGUCCGCCAGAUAUA----UGGAAAUGAAAUGUUUUACGACCGGACUGCACUUUUUUCG ((((((((((((..((((((((((((......)))))).............-------....))))))..))))))----)))))).(((((((....(....)....)))....)))). ( -26.52) >consensus UUUUUAUGUGUCGCGUGGACGCGACAAGCUGCUGUCGUAGCUUUUUUUU_________UAUUGUCCGCCAGAUAUA____UGGA_____AAUGUUUUACGACCGGACUGCACUUUUUUCG ......(((((((((....))))))(((((((....)))))))...................((((((((..........))).........(((....)))))))).)))......... (-22.16 = -22.73 + 0.56)

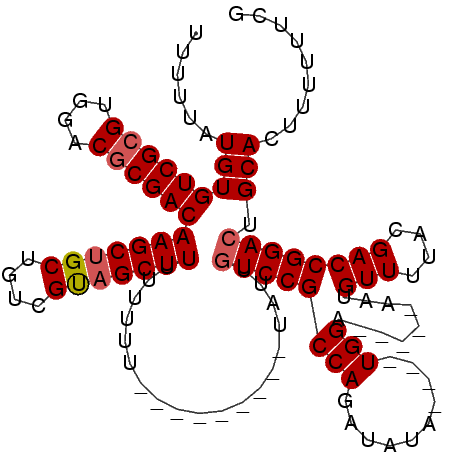
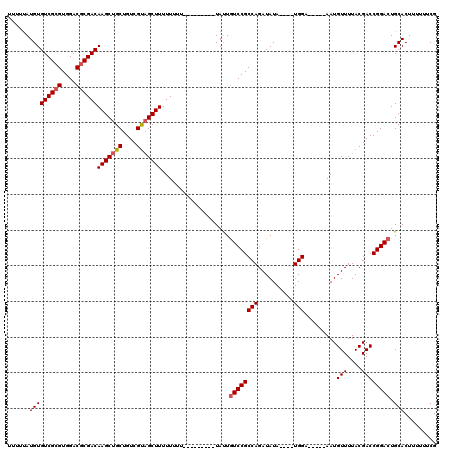
| Location | 16,702,752 – 16,702,868 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.93 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -23.81 |
| Energy contribution | -24.38 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16702752 116 + 22407834 ----UAUAUCUGGCGGACAGUAAAUAAUAAAAAAAAAAAGCUACGACAGCAGCUUGUCGAGUCCACGCGACACAUGAAAAUAUUGCUUUGUUUUUUCCGCUUUGUAUUUCGGGGAUUGCU ----.((((..((((((.............(((((.(((((..((((((....)))))).(((.....))).............))))).)))))))))))..))))............. ( -22.91) >DroSec_CAF1 157441 107 + 1 ----UAUAUCUGGCGGACAAUA---------AAAAAAAAGCUACGACAGCAGCUUGUCGCGUCCACGCGACACAUAAAAAUAUUGCUUUGUUUUUUCCGGUUUGUAUUUCGGGGAUCGCU ----.......(((((.(((((---------......(((((.(....).)))))((((((....)))))).........)))))........(..((((........))))..)))))) ( -29.20) >DroSim_CAF1 157371 111 + 1 GAUGUAUAUCUGGCGGAUAAUA---------AAAAAAAAGCUACGACAGCAGCUUGUCGCGUCCACGCGACACAUAAAAAUAUUGCUUUGUUUUUUCCGCUUUGUAUUUCGGGGAUCGCU ((.(((((...((((((.((((---------((....(((((.(....).)))))((((((....))))))...............))))))...)))))).))))).)).......... ( -31.90) >DroEre_CAF1 158839 109 + 1 ----UAUAUCUGGCGGACAAUA-------AUAAAAAAAAGCAGCGACAGCAGCUUGUCGCGUCCACGCGACACAUAAAAAUAUUGCUUUGUUUUUUCCGCUUUGUAUUUCGGGGAUCGCA ----.((((..((((((....(-------(((((...((((.((....)).))))((((((....))))))...............))))))...))))))..))))............. ( -29.70) >consensus ____UAUAUCUGGCGGACAAUA_________AAAAAAAAGCUACGACAGCAGCUUGUCGCGUCCACGCGACACAUAAAAAUAUUGCUUUGUUUUUUCCGCUUUGUAUUUCGGGGAUCGCU .....((((..((((((....................(((((.(....).)))))((((((....))))))....(((((((......)))))))))))))..))))............. (-23.81 = -24.38 + 0.56)


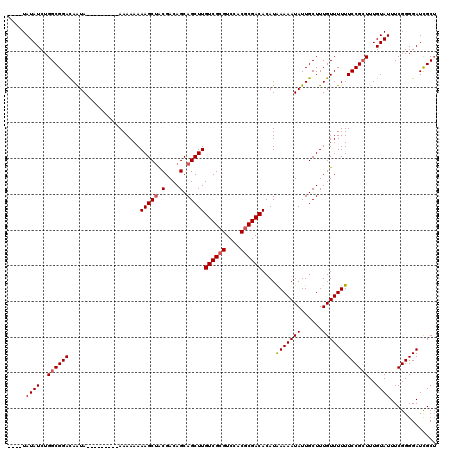
| Location | 16,702,828 – 16,702,947 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.40 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -20.26 |
| Energy contribution | -21.07 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16702828 119 - 22407834 GCGUAUUUUGAGCGGUGACAAGAUUGCUUACAAAACACACACAAAGUGAAAAA-GUAUUUGUCAAGAAGCUAGAAGGAAAAGCAAUCCCCGAAAUACAAAGCGGAAAAAACAAAGCAAUA ((((((((((...(....)..((((((((........(((.....)))....(-((.(((....))).)))........))))))))..)))))))).....(.......)...)).... ( -23.30) >DroSec_CAF1 157508 119 - 1 GCGUAUUUUGAGCGGUGACAAGAUUGCUUACAAAACACACACAAAGUGAAAAA-GUAUUUGGCAAGAAGCUAGAAGGAAAAGCGAUCCCCGAAAUACAAACCGGAAAAAACAAAGCAAUA ((((((((((...(....)..((((((((........(((.....))).....-...((((((.....)))))).....))))))))..)))))))).....(.......)...)).... ( -26.80) >DroSim_CAF1 157442 119 - 1 GCGUAUUUUGAGCGGUGACAAGAUUGGUUACAAAACACACACAAAGUGAAAAA-GUAUUUGGCAAGAAGCUAGAAGGAAAAGCGAUCCCCGAAAUACAAAGCGGAAAAAACAAAGCAAUA ((((((((((...(....)..(((((.((........(((.....))).....-...((((((.....)))))).....)).)))))..)))))))).....(.......)...)).... ( -22.30) >DroEre_CAF1 158908 118 - 1 GCGUAUUUUGAGCGCUGACAAGAUUGCUUACAAAACA--CACAAAGUGAAAAACACAUUUGGCAAGAAGCUAGAAGGAAAUGCGAUCCCCGAAAUACAAAGCGGAAAAAACAAAGCAAUA ((((((((((.(......)..((((((........((--(.....))).......(.((((((.....)))))).).....))))))..)))))))).....(.......)...)).... ( -20.40) >consensus GCGUAUUUUGAGCGGUGACAAGAUUGCUUACAAAACACACACAAAGUGAAAAA_GUAUUUGGCAAGAAGCUAGAAGGAAAAGCGAUCCCCGAAAUACAAAGCGGAAAAAACAAAGCAAUA ((((((((((...(....)..((((((((......(((.......))).........((((((.....)))))).....))))))))..)))))))).....(.......)...)).... (-20.26 = -21.07 + 0.81)

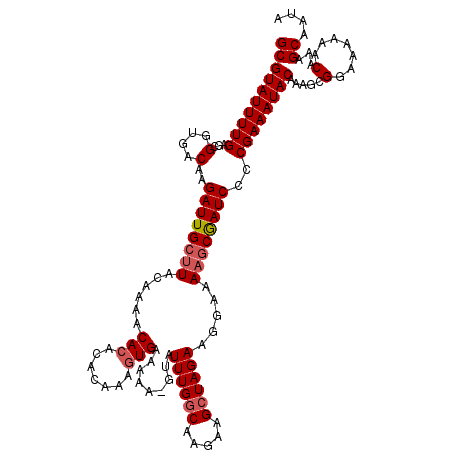
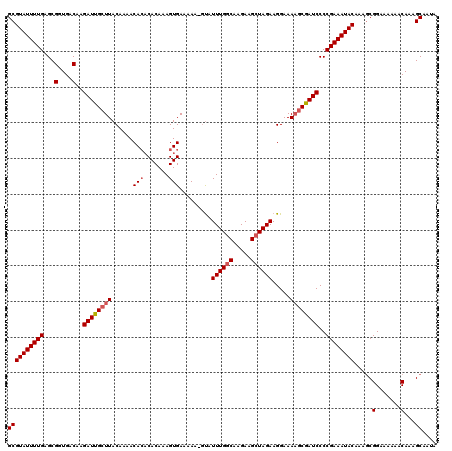
| Location | 16,702,907 – 16,703,027 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.35 |
| Mean single sequence MFE | -30.01 |
| Consensus MFE | -24.33 |
| Energy contribution | -24.83 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16702907 120 + 22407834 GUGUGUUUUGUAAGCAAUCUUGUCACCGCUCAAAAUACGCCUGCGCAUUUGCCUGGCUCAUUUUAAUGCUUAGAUCAUGCAGAUAGCCAGGCCAACGAACAUUUCAAUGUACAUACAGUG ((((((((((..(((............))))))))))))).((..((((.((((((((.(((....(((.........))))))))))))))....((.....))))))..))....... ( -30.70) >DroSec_CAF1 157587 108 + 1 GUGUGUUUUGUAAGCAAUCUUGUCACCGCUCAAAAUACGCCUGCGCAUUUGCCUGGCUCAUUAUAAUGCUUAGAUCAUGCAGAUAGCCAGGCCAACG------------UAAGUACAGUG ((((((((((..(((............)))))))))))))(((.......((((((((.(((....(((.........))))))))))))))..((.------------...)).))).. ( -30.70) >DroSim_CAF1 157521 108 + 1 GUGUGUUUUGUAACCAAUCUUGUCACCGCUCAAAAUACGCCUGCGCAUUUGCCUGGCUCAUUAUAAUGCUUAGAUCAUGCAGAUAGCCAGGCCAACG------------UAAGUACAGUG ((((((((((....................))))))))))(((.......((((((((.(((....(((.........))))))))))))))..((.------------...)).))).. ( -27.95) >DroEre_CAF1 158988 114 + 1 G--UGUUUUGUAAGCAAUCUUGUCAGCGCUCAAAAUACGCCUGCGCAUUUGCCUGGCUCAUUAUAAUGCCCAGAUCAUGCAGAUAGCCAGGCCAACGAAUAUUUCAA----UGUACAGUG (--(((((((...((..........))...))))))))..(((..((((.((((((((.(((...(((.......)))...)))))))))))....((.....))))----))..))).. ( -30.70) >consensus GUGUGUUUUGUAAGCAAUCUUGUCACCGCUCAAAAUACGCCUGCGCAUUUGCCUGGCUCAUUAUAAUGCUUAGAUCAUGCAGAUAGCCAGGCCAACG____________UAAGUACAGUG ..((((((((..(((............)))))))))))(((((.((((((((.((((((((....)))...)).))).)))))).)))))))............................ (-24.33 = -24.83 + 0.50)

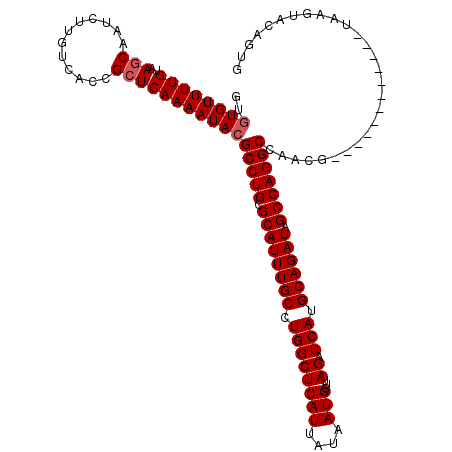
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:31 2006