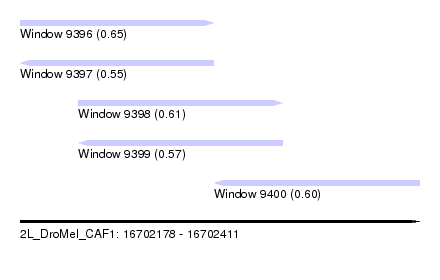
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,702,178 – 16,702,411 |
| Length | 233 |
| Max. P | 0.648175 |

| Location | 16,702,178 – 16,702,291 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -35.95 |
| Consensus MFE | -25.61 |
| Energy contribution | -26.99 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16702178 113 + 22407834 UUGUUUGGCAAC------GGCAGUGCGCAGCUCUGAUCGAAAUGCUCCAUUGGGUGCACACACCCAGCCCAACAAGGCAUUG-GUUUAGCCCACCCAUCCAAAGCCGAUGACCUUGCCCA ..(((.(((..(------(((((.((...)).))).)))...........((((((....))))))))).)))(((((((((-((((..............))))))))).))))..... ( -36.34) >DroSec_CAF1 156898 112 + 1 UUGUUUGGCAACA----CG---GUGCGCAGCUCUGAUCGAAAUGCUCCAUUGGGUGCACACACCCAGCCCAACAAGGCAUUGGGUUCAGCCCACCCAUCCA-AGCCGAUGACCUUGCCCA (((((.(((....----.(---(...(((..((.....))..))).))..((((((....))))))))).)))))((((.((((.....))))..((((..-....))))....)))).. ( -37.40) >DroSim_CAF1 156842 114 + 1 UUGUUUGGCAACA----CGGCAGUGCGCAGCUCUGAUCGAAAUGCUCCAUUGGUUGCACACACCCAGCCCAACAAGGCAUUG-GUUCAGCCCACCCAUCCA-AGCCGAUGACCUUGCCCA ......(((((((----((((.(((.((((((..(((.((.....)).))))))))).)))(((..(((......)))...)-))................-.)))).))...))))).. ( -31.20) >DroEre_CAF1 158297 119 + 1 UUGCUUAGCAGCAAAAAAGGCAGUGCGCAGCUCUAAUCGAAAUGCUCCAAUGGGUGCACACACCCAGCCCAAAAAGGCAUUG-GUUCAAACCACCCAUCCAGAGCCGAUGACCUUGCCCA (((((....)))))....(((.(.(((((..((.....))..))).....((((((....)))))))).)...(((((((((-((((..............))))))))).))))))).. ( -38.84) >consensus UUGUUUGGCAACA____CGGCAGUGCGCAGCUCUGAUCGAAAUGCUCCAUUGGGUGCACACACCCAGCCCAACAAGGCAUUG_GUUCAGCCCACCCAUCCA_AGCCGAUGACCUUGCCCA ......(((.........(((.....(((..((.....))..))).....((((((....)))))))))....(((((((((.((((..............))))))))).))))))).. (-25.61 = -26.99 + 1.38)

| Location | 16,702,178 – 16,702,291 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -43.57 |
| Consensus MFE | -33.31 |
| Energy contribution | -34.25 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16702178 113 - 22407834 UGGGCAAGGUCAUCGGCUUUGGAUGGGUGGGCUAAAC-CAAUGCCUUGUUGGGCUGGGUGUGUGCACCCAAUGGAGCAUUUCGAUCAGAGCUGCGCACUGCC------GUUGCCAAACAA ..((((((((...(((((((((.((((((.((...((-(...((((....))))..)))..)).)))))).((((....)))).)))))))))......)))------.)))))...... ( -45.30) >DroSec_CAF1 156898 112 - 1 UGGGCAAGGUCAUCGGCU-UGGAUGGGUGGGCUGAACCCAAUGCCUUGUUGGGCUGGGUGUGUGCACCCAAUGGAGCAUUUCGAUCAGAGCUGCGCAC---CG----UGUUGCCAAACAA .((((.((.(((((((((-(.(.((((((.((...(((((..((((....)))))))))..)).)))))).).))))....))))..)).))..)).)---).----((((....)))). ( -44.90) >DroSim_CAF1 156842 114 - 1 UGGGCAAGGUCAUCGGCU-UGGAUGGGUGGGCUGAAC-CAAUGCCUUGUUGGGCUGGGUGUGUGCAACCAAUGGAGCAUUUCGAUCAGAGCUGCGCACUGCCG----UGUUGCCAAACAA ..(((((.(.(..(((((-..(..((.(((......)-))...))...)..)))))((((.((((..((...)).(((.(((.....))).))))))))))))----).)))))...... ( -38.50) >DroEre_CAF1 158297 119 - 1 UGGGCAAGGUCAUCGGCUCUGGAUGGGUGGUUUGAAC-CAAUGCCUUUUUGGGCUGGGUGUGUGCACCCAUUGGAGCAUUUCGAUUAGAGCUGCGCACUGCCUUUUUUGCUGCUAAGCAA .(((((..((...(((((((((.....(((..((..(-((((((((....)))).(((((....))))))))))..))..))).))))))))).))..)))))...(((((....))))) ( -45.60) >consensus UGGGCAAGGUCAUCGGCU_UGGAUGGGUGGGCUGAAC_CAAUGCCUUGUUGGGCUGGGUGUGUGCACCCAAUGGAGCAUUUCGAUCAGAGCUGCGCACUGCCG____UGUUGCCAAACAA ..((((..((...(((((((((.((((...(((.........((((....))))((((((....))))))....)))..)))).))))))))).))..))))......(((....))).. (-33.31 = -34.25 + 0.94)

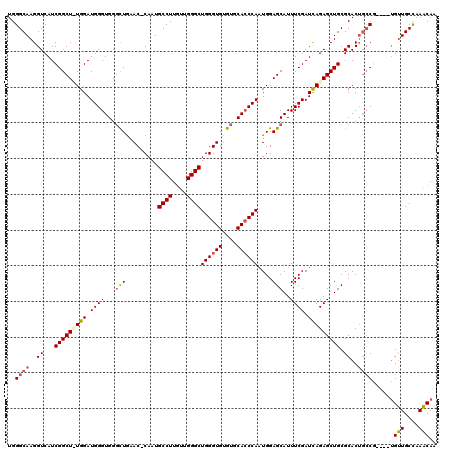
| Location | 16,702,212 – 16,702,331 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.85 |
| Mean single sequence MFE | -34.64 |
| Consensus MFE | -25.92 |
| Energy contribution | -27.04 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16702212 119 + 22407834 AAUGCUCCAUUGGGUGCACACACCCAGCCCAACAAGGCAUUG-GUUUAGCCCACCCAUCCAAAGCCGAUGACCUUGCCCAUAUCCACUGAAGUAUAUACACGGGUGGUAUGCCCAAAAAG .........((((((..((.(((((.......((((((((((-((((..............))))))))).)))))...((((.(......).))))....)))))))..)))))).... ( -36.24) >DroSec_CAF1 156931 119 + 1 AAUGCUCCAUUGGGUGCACACACCCAGCCCAACAAGGCAUUGGGUUCAGCCCACCCAUCCA-AGCCGAUGACCUUGCCCAUAUUCACUGAAGUAUACACACGGGUGGUAUGCCCAAAAAG ..........((((((....))))))(((......))).((((((.....((((((...((-((........))))...(((((......)))))......))))))...)))))).... ( -34.80) >DroSim_CAF1 156878 118 + 1 AAUGCUCCAUUGGUUGCACACACCCAGCCCAACAAGGCAUUG-GUUCAGCCCACCCAUCCA-AGCCGAUGACCUUGCCCAUAUCCACUGAAGUAUACACACGGGUGGUAUGCCCAAAAAG .........((((..(((..(((((.......((((((((((-(((...............-)))))))).)))))...............((......)))))))...))))))).... ( -29.46) >DroEre_CAF1 158337 119 + 1 AAUGCUCCAAUGGGUGCACACACCCAGCCCAAAAAGGCAUUG-GUUCAAACCACCCAUCCAGAGCCGAUGACCUUGCCCAUAUGCACUGGAGUAUAUACACGAGUGGCAUGCCCAAAAAG .((((((((.((((((....))))))((.....(((((((((-((((..............))))))))).))))........))..))))))))........(.((....)))...... ( -38.06) >consensus AAUGCUCCAUUGGGUGCACACACCCAGCCCAACAAGGCAUUG_GUUCAGCCCACCCAUCCA_AGCCGAUGACCUUGCCCAUAUCCACUGAAGUAUACACACGGGUGGUAUGCCCAAAAAG .(((((.((.((((((....))))))......((((((((((.((((..............))))))))).)))))...........)).)))))......((((.....))))...... (-25.92 = -27.04 + 1.12)

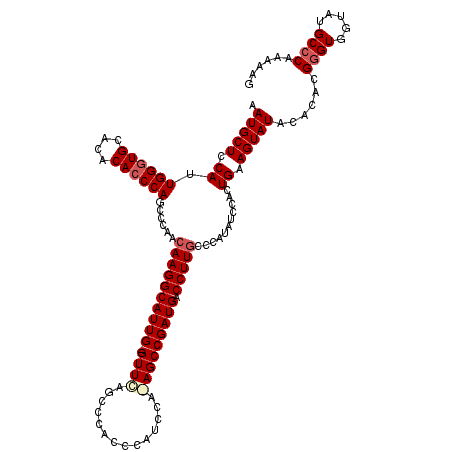
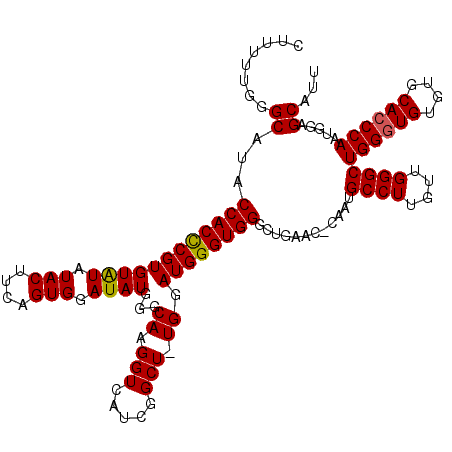
| Location | 16,702,212 – 16,702,331 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.85 |
| Mean single sequence MFE | -43.90 |
| Consensus MFE | -34.16 |
| Energy contribution | -33.98 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16702212 119 - 22407834 CUUUUUGGGCAUACCACCCGUGUAUAUACUUCAGUGGAUAUGGGCAAGGUCAUCGGCUUUGGAUGGGUGGGCUAAAC-CAAUGCCUUGUUGGGCUGGGUGUGUGCACCCAAUGGAGCAUU ........((...((((((((((...(((....))).)))))))((((((.....))))))..((((((.((...((-(...((((....))))..)))..)).)))))).))).))... ( -43.90) >DroSec_CAF1 156931 119 - 1 CUUUUUGGGCAUACCACCCGUGUGUAUACUUCAGUGAAUAUGGGCAAGGUCAUCGGCU-UGGAUGGGUGGGCUGAACCCAAUGCCUUGUUGGGCUGGGUGUGUGCACCCAAUGGAGCAUU .....(((.....)))(((((((...(((....))).)))))))...........(((-(.(.((((((.((...(((((..((((....)))))))))..)).)))))).).))))... ( -45.50) >DroSim_CAF1 156878 118 - 1 CUUUUUGGGCAUACCACCCGUGUGUAUACUUCAGUGGAUAUGGGCAAGGUCAUCGGCU-UGGAUGGGUGGGCUGAAC-CAAUGCCUUGUUGGGCUGGGUGUGUGCAACCAAUGGAGCAUU ....(((.(((((((.(((((((.(((......))).))))))).........(((((-..(..((.(((......)-))...))...)..)))))))))))).)))............. ( -39.90) >DroEre_CAF1 158337 119 - 1 CUUUUUGGGCAUGCCACUCGUGUAUAUACUCCAGUGCAUAUGGGCAAGGUCAUCGGCUCUGGAUGGGUGGUUUGAAC-CAAUGCCUUUUUGGGCUGGGUGUGUGCACCCAUUGGAGCAUU ........((...(((...((((((((((.(((((.((...(((((.(((((.(.((((.....)))).)..)).))-)..)))))...)).)))))))))))))))....))).))... ( -46.30) >consensus CUUUUUGGGCAUACCACCCGUGUAUAUACUUCAGUGGAUAUGGGCAAGGUCAUCGGCU_UGGAUGGGUGGGCUGAAC_CAAUGCCUUGUUGGGCUGGGUGUGUGCACCCAAUGGAGCAUU ........((...((((((((((((.(((....))).))))...((.(((.....))).)).))))))))............((((....))))((((((....)))))).....))... (-34.16 = -33.98 + -0.19)

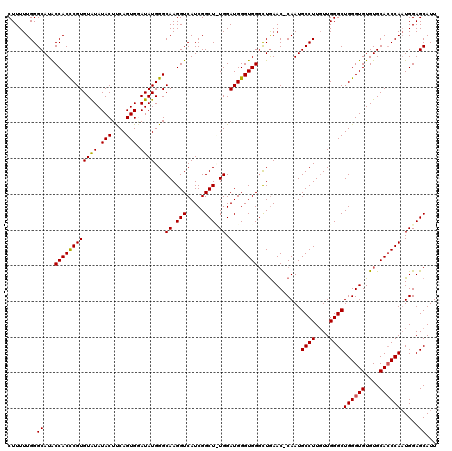
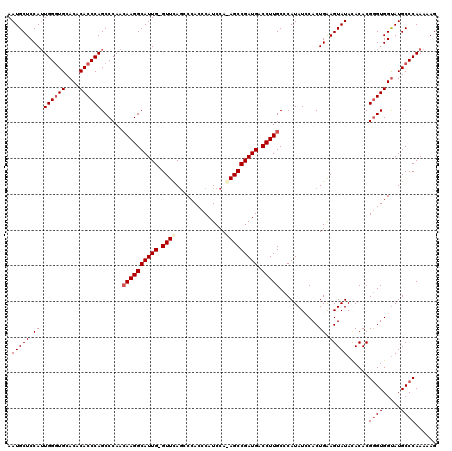
| Location | 16,702,291 – 16,702,411 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.39 |
| Mean single sequence MFE | -35.35 |
| Consensus MFE | -21.45 |
| Energy contribution | -22.20 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16702291 120 - 22407834 UUGUCAUUGAAAUGCUCAGAUAUGUCUGGCUUCAGCCAGAUCUGGAAAAACUGGAGCAUGCUACGUUUCGGUUUGGGCCUCUUUUUGGGCAUACCACCCGUGUAUAUACUUCAGUGGAUA ...(((((((((((((((((...(((((((....)))))))..((..((((((((((.......))))))))))...))....))))))))).......(((....)))))))))))... ( -40.40) >DroSec_CAF1 157010 105 - 1 UUGUCAUUGAAAUGC---------------UUCAGCCAGAUCUGGAAAAACUGGAGCAUGCUACGUUUCGAUUGGGGCCUCUUUUUGGGCAUACCACCCGUGUGUAUACUUCAGUGAAUA ...((((((((((((---------------((((((((....))).....)))))))))..((((...((..(((.((((......))))...)))..))..))))...))))))))... ( -33.40) >DroSim_CAF1 156956 105 - 1 UUGUCAUUGAAAUGC---------------UUCAGCCAGAUCUGGAAAAACUGGAGCAUGCUACGUUUCGAUUGGGGCCUCUUUUUGGGCAUACCACCCGUGUGUAUACUUCAGUGGAUA ...((((((((((((---------------((((((((....))).....)))))))))..((((...((..(((.((((......))))...)))..))..))))...))))))))... ( -32.50) >DroEre_CAF1 158416 117 - 1 UUGUCAUUGAAAUGCUCUCUGAUGUCUGGCUGUAGCCAGAUCUGGAAAAACUGG---AUGCUACGUUUCGAUUGGGGCCUCUUUUUGGGCAUGCCACUCGUGUAUAUACUCCAGUGCAUA ...........((((..(((...(((((((....)))))))..)))...(((((---(...((((...(((.(((.((((......))))...))).))))))).....)))))))))). ( -35.10) >consensus UUGUCAUUGAAAUGC_______________UUCAGCCAGAUCUGGAAAAACUGGAGCAUGCUACGUUUCGAUUGGGGCCUCUUUUUGGGCAUACCACCCGUGUAUAUACUUCAGUGGAUA ..............................(((..(((....)))....(((((((.((((.(((...(((.(((.((((......))))...))).)))))))))).)))))))))).. (-21.45 = -22.20 + 0.75)

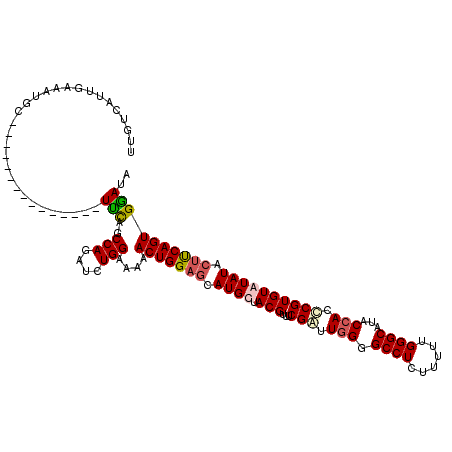
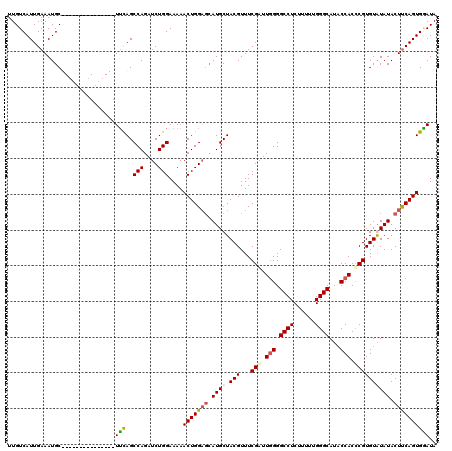
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:24 2006