
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,694,695 – 16,694,831 |
| Length | 136 |
| Max. P | 0.980910 |

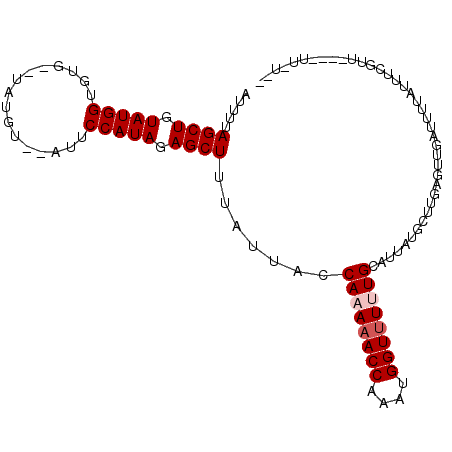
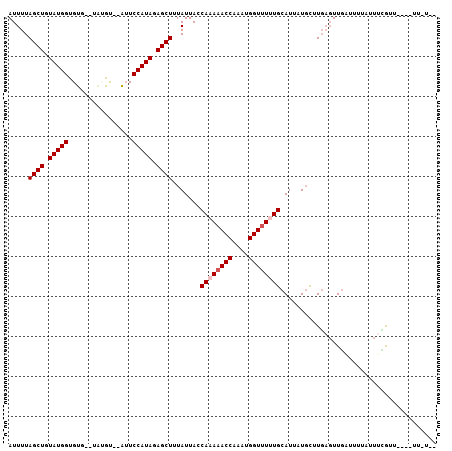
| Location | 16,694,695 – 16,694,798 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.62 |
| Mean single sequence MFE | -20.48 |
| Consensus MFE | -16.79 |
| Energy contribution | -17.29 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16694695 103 + 22407834 AUUUUAGCUGUAUGGUGUU--UAUGUAUAUUCCAUAGAGCUUUAUUACCAAAAACCAAAUGGUUUCUGCAUUAUGUUUGAGUUGAUUUUAUUUGGUAUU-CUU-UAC .....((((.(((((.((.--.......)).))))).))))............(((((((((..((.((.(((....))))).))..)))))))))...-...-... ( -22.90) >DroEre_CAF1 149877 102 + 1 AUUUUAGCUGUAUGGUGUG--UAUGU--GUUCCAUAGAGCUUUAUUACCAAAAACCAAAUGGUUUUUGCAUUAUGCUUGAGUUGAUUUUUUUUCGUUUU-GUUACAU ...((((((((((((((.(--((...--((((....)))).....)))((((((((....))))))))))))))))...))))))..............-....... ( -20.60) >DroWil_CAF1 252464 104 + 1 AUUUUAGCUGUAUGGUGUA--UGAGCAUAAUCCAUAGAGCUUUAUUACCAAAAACCAAAUGGUUUUUGCAUUUUGCUUGAGUUGUUU-UCUUUCGUAUUGAUUUUGA .....((((.(((((..((--(....)))..))))).)))).......((((((((....))))))))((...(((..(((......-.)))..))).))....... ( -22.00) >DroYak_CAF1 149260 95 + 1 AUUUUAGCUGUAUGGUGUGUUUAUGU--GUUCCAUAGAGCUUUAUUACCAAAAACCAAAUGGUUUCUGCAUUAUGCUUGAGUUGAUUUUUUUUCAUU---------- ...(((((((((((((((.((((((.--....)))))).............(((((....)))))..)))))))))...))))))............---------- ( -17.70) >DroMoj_CAF1 76423 91 + 1 AUUUUAGCUGUAUGGUGUG--CGCGU--AUUCCAUAGAGCUUUAUUACCAAAAACCAAAUGGUCUUUGCAU--UGCUUUCUUUUCGUUUAUUCAGUU---------- .....((((.(((((.(((--....)--)).))))).)))).......((((.(((....))).))))...--........................---------- ( -17.40) >DroAna_CAF1 89532 97 + 1 AUUUUAGCUGUAUGGUGUC--UC-AU--AUUCCAUAGAGCUUUAUUACCAAAAACCAAAUGGUUUUUGCAUUUUUUCUGAGUUGCUUUUGUUGUG-----GUUAUGA .....((((.(((((....--..-..--...))))).))))((((.((((((((((....)))))))(((.(((....))).))).........)-----)).)))) ( -22.30) >consensus AUUUUAGCUGUAUGGUGUG__UAUGU__AUUCCAUAGAGCUUUAUUACCAAAAACCAAAUGGUUUUUGCAUUAUGCUUGAGUUGAUUUUAUUUCGUU____UU_U__ .....((((.(((((................))))).)))).......((((((((....))))))))....................................... (-16.79 = -17.29 + 0.50)

| Location | 16,694,695 – 16,694,798 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.62 |
| Mean single sequence MFE | -18.08 |
| Consensus MFE | -17.60 |
| Energy contribution | -17.24 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
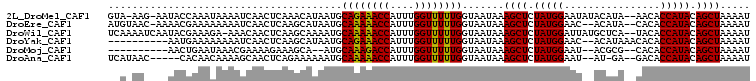
>2L_DroMel_CAF1 16694695 103 - 22407834 GUA-AAG-AAUACCAAAUAAAAUCAACUCAAACAUAAUGCAGAAACCAUUUGGUUUUUGGUAAUAAAGCUCUAUGGAAUAUACAUA--AACACCAUACAGCUAAAAU ...-...-...............................((((((((....)))))))).......((((.(((((..........--....))))).))))..... ( -17.44) >DroEre_CAF1 149877 102 - 1 AUGUAAC-AAAACGAAAAAAAAUCAACUCAAGCAUAAUGCAAAAACCAUUUGGUUUUUGGUAAUAAAGCUCUAUGGAAC--ACAUA--CACACCAUACAGCUAAAAU .......-...............................((((((((....)))))))).......((((.(((((...--.....--....))))).))))..... ( -17.50) >DroWil_CAF1 252464 104 - 1 UCAAAAUCAAUACGAAAGA-AAACAACUCAAGCAAAAUGCAAAAACCAUUUGGUUUUUGGUAAUAAAGCUCUAUGGAUUAUGCUCA--UACACCAUACAGCUAAAAU ............(....).-...................((((((((....)))))))).......((((.(((((..(((....)--))..))))).))))..... ( -19.90) >DroYak_CAF1 149260 95 - 1 ----------AAUGAAAAAAAAUCAACUCAAGCAUAAUGCAGAAACCAUUUGGUUUUUGGUAAUAAAGCUCUAUGGAAC--ACAUAAACACACCAUACAGCUAAAAU ----------.............................((((((((....)))))))).......((((.(((((...--...........))))).))))..... ( -17.44) >DroMoj_CAF1 76423 91 - 1 ----------AACUGAAUAAACGAAAAGAAAGCA--AUGCAAAGACCAUUUGGUUUUUGGUAAUAAAGCUCUAUGGAAU--ACGCG--CACACCAUACAGCUAAAAU ----------........................--...((((((((....)))))))).......((((.(((((...--.....--....))))).))))..... ( -17.60) >DroAna_CAF1 89532 97 - 1 UCAUAAC-----CACAACAAAAGCAACUCAGAAAAAAUGCAAAAACCAUUUGGUUUUUGGUAAUAAAGCUCUAUGGAAU--AU-GA--GACACCAUACAGCUAAAAU .....((-----(.........(((............)))(((((((....)))))))))).....((((.(((((...--..-..--....))))).))))..... ( -18.60) >consensus __A_AA____AACGAAAAAAAAUCAACUCAAGCAUAAUGCAAAAACCAUUUGGUUUUUGGUAAUAAAGCUCUAUGGAAU__ACAUA__CACACCAUACAGCUAAAAU .......................................((((((((....)))))))).......((((.(((((................))))).))))..... (-17.60 = -17.24 + -0.36)
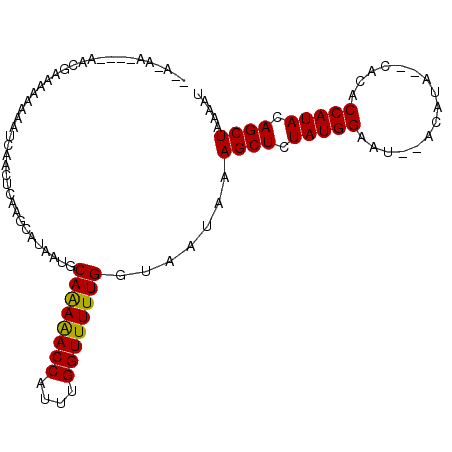
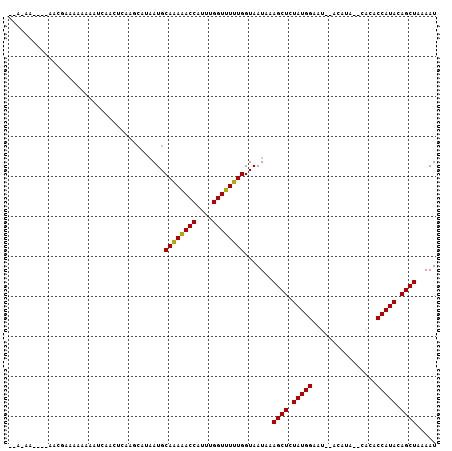
| Location | 16,694,727 – 16,694,831 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 72.51 |
| Mean single sequence MFE | -11.95 |
| Consensus MFE | -8.52 |
| Energy contribution | -8.43 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16694727 104 - 22407834 AAAAUGACAUUGAGAACAAGUAACGCGAGAAAAGUA-AAGAAUACCAAAUAAAAUCAACUCAAAC---AUAAUGCAGAAACCAUUUGGUUUUUGGUAAUAAAGCUCUA .........(((((.........(....)....(((-.....))).............)))))..---......((((((((....)))))))).............. ( -14.00) >DroSec_CAF1 149496 104 - 1 AUAAUAACAUUAAGAAAAAGUAACACUAGAAAAGUA-AAUUAUACCAAAUAAAAUCAACUCAAGC---AUAAUGCAGAAACCAUUUGGUUUUUGGUAAUAAAGCUCUA ............(((.........(((.....))).-.....((((.................((---.....))(((((((....))))))))))).......))). ( -12.40) >DroEre_CAF1 149907 105 - 1 ACAAUAAGAUUAAGAACAAGUAACGUUAGAAAAAUGUAACAAAACGAAAAAAAAUCAACUCAAGC---AUAAUGCAAAAACCAUUUGGUUUUUGGUAAUAAAGCUCUA ............(((.(..((.(((((.....))))).)).........................---......((((((((....))))))))........).))). ( -13.30) >DroYak_CAF1 149292 94 - 1 CUAAUACCAUUAAAAACAAAUAAAGUUAGAA-----------AAUGAAAAAAAAUCAACUCAAGC---AUAAUGCAGAAACCAUUUGGUUUUUGGUAAUAAAGCUCUA ....((((................(((.((.-----------..(((.......)))..)).)))---.......(((((((....)))))))))))........... ( -11.90) >DroAna_CAF1 89561 95 - 1 AAAACAAAGU-----AUAAA-AAUCUUAAAAAAUCAUAAC----CACAACAAAAGCAACUCAGAA---AAAAUGCAAAAACCAUUUGGUUUUUGGUAAUAAAGCUCUA ..........-----.....-.................((----(.........(((........---....)))(((((((....))))))))))............ ( -9.90) >DroPer_CAF1 169704 87 - 1 ---AU---UUUGUUGACAAAAAACAUUUUAA---------------AAACAUAAGCAACUCAAGCGAAAAAAUGCAAAAACCAUUUGGGUUUUGGUGAUAAAGCUCUA ---.(---((((((..(((((..(.......---------------........((.......))....(((((.......))))))..)))))..)))))))..... ( -10.20) >consensus AAAAUAACAUUAAGAACAAAUAACGUUAGAAAA_UA_AA____ACCAAAAAAAAUCAACUCAAGC___AUAAUGCAAAAACCAUUUGGUUUUUGGUAAUAAAGCUCUA ..........................................................................((((((((....)))))))).............. ( -8.52 = -8.43 + -0.08)

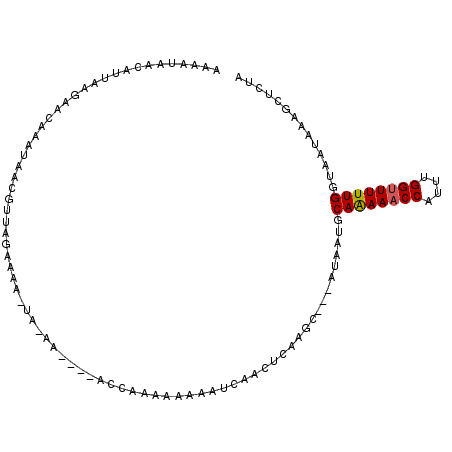
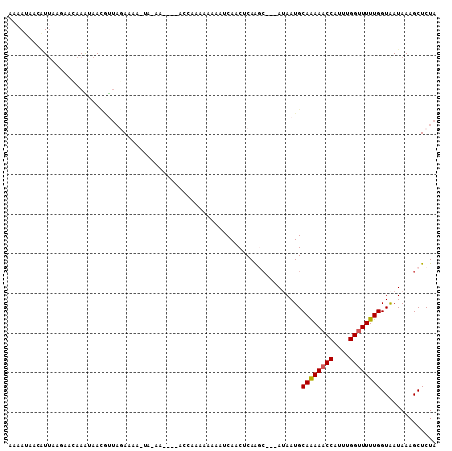
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:15 2006