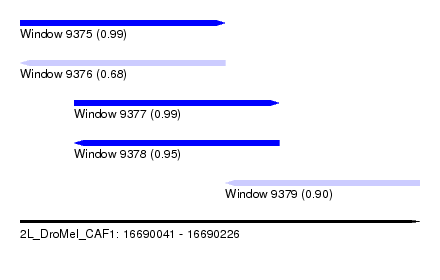
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,690,041 – 16,690,226 |
| Length | 185 |
| Max. P | 0.992259 |

| Location | 16,690,041 – 16,690,136 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 73.76 |
| Mean single sequence MFE | -30.85 |
| Consensus MFE | -17.86 |
| Energy contribution | -18.37 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
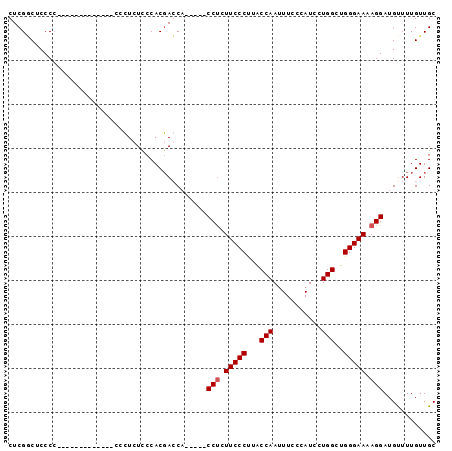
>2L_DroMel_CAF1 16690041 95 + 22407834 AGCGGUCUAGCGCCUG--UUGGGUUUGGCAACAAAACAUCCUUUUCCCAGCCACGAUGGGAAAUUGGUAAGGGAAGGGG---UGGGGUGGUCGUGGAAUA .(((..(....((((.--.....((((....)))).(((((((((((..((((...........))))..)))))))))---)))))))..)))...... ( -32.40) >DroSec_CAF1 144791 89 + 1 AGCGGUCUAGCGCCUG--UUG-GUUGGGCAACAAAAUAUCCUUUUCCCAGCCAGGAUAUUAAAUUGGCAAGGGAAGAGG--------UGUUCGUGGAAGA .((((((((((.....--...-))))))).....((((.(((((((((.(((((.........)))))..)))))))))--------)))))))...... ( -30.40) >DroSim_CAF1 144702 89 + 1 AGCGGUCUAGCGCCUG--UUG-GUUGGGCAACAAAACAUCCUUUUCCCAGCCAGGAUAUUAAAUUGGUAAGGGAAGAGG--------UGGUAGUGGAAGA .((......))(((((--...-..)))))..((..((..(((((((((.(((((.........)))))..)))))))))--------..))..))..... ( -28.80) >DroEre_CAF1 145203 91 + 1 AGCGGUCUAGCGCCUGUGUUG-GUUGGGCAACAAAGCAUCCUUUUCCCAGCCAGGAUGGCAAGUUGGCAGGGGAAGAGG--------AGCUCGUGGGAGA .((((((((..(((......)-))))))).....(((.((((((((((.(((((.........)))))..)))))))))--------)))))))...... ( -34.60) >DroWil_CAF1 245856 83 + 1 ----GCCUGGCGCCUG--UUG-GGUAUGCAGCAAAAAAAAUCUCUCUAAGCUGC---------UUGGAAAAGGAGGUUAGGUUGGGUUGGUUGUGUUGU- ----(((..((....)--)..-)))..((((((.((..(((((.(((((.((.(---------((....))).)).)))))..)))))..)).))))))- ( -28.10) >DroYak_CAF1 144516 89 + 1 AGCGGUCUAGCGCCUG--UUG-GUUUGGCAACAAAACAUCCUUUUCCCAGCCAGGAUAGGAAAUUGGUAAGGGAAGAGG--------AGCUCGUGGGAGA .((......)).((..--(.(-(((((....)))....((((((((((.(((((.........)))))..)))))))))--------)))).)..))... ( -30.80) >consensus AGCGGUCUAGCGCCUG__UUG_GUUGGGCAACAAAACAUCCUUUUCCCAGCCAGGAUAGGAAAUUGGUAAGGGAAGAGG________UGGUCGUGGAAGA ....(((((((...........)))))))..........(((((((((.(((((.........)))))..)))))))))..................... (-17.86 = -18.37 + 0.51)

| Location | 16,690,041 – 16,690,136 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 73.76 |
| Mean single sequence MFE | -20.22 |
| Consensus MFE | -11.24 |
| Energy contribution | -12.18 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
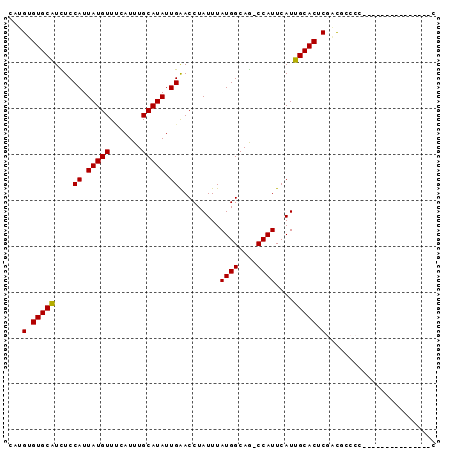
>2L_DroMel_CAF1 16690041 95 - 22407834 UAUUCCACGACCACCCCA---CCCCUUCCCUUACCAAUUUCCCAUCGUGGCUGGGAAAAGGAUGUUUUGUUGCCAAACCCAA--CAGGCGCUAGACCGCU .......((.((......---((..(((((...(((...........)))..)))))..))......(((((.......)))--)))))).......... ( -16.60) >DroSec_CAF1 144791 89 - 1 UCUUCCACGAACA--------CCUCUUCCCUUGCCAAUUUAAUAUCCUGGCUGGGAAAAGGAUAUUUUGUUGCCCAAC-CAA--CAGGCGCUAGACCGCU .............--------(((.(((((..((((...........)))).))))).)))......(((((......-)))--))((((......)))) ( -22.80) >DroSim_CAF1 144702 89 - 1 UCUUCCACUACCA--------CCUCUUCCCUUACCAAUUUAAUAUCCUGGCUGGGAAAAGGAUGUUUUGUUGCCCAAC-CAA--CAGGCGCUAGACCGCU .............--------(((.(((((...(((...........)))..))))).)))......(((((......-)))--))((((......)))) ( -19.00) >DroEre_CAF1 145203 91 - 1 UCUCCCACGAGCU--------CCUCUUCCCCUGCCAACUUGCCAUCCUGGCUGGGAAAAGGAUGCUUUGUUGCCCAAC-CAACACAGGCGCUAGACCGCU ........(((((--------(((.(((((..((((...........)))).))))).)))).))))(((((......-)))))..((((......)))) ( -28.20) >DroWil_CAF1 245856 83 - 1 -ACAACACAACCAACCCAACCUAACCUCCUUUUCCAA---------GCAGCUUAGAGAGAUUUUUUUUGCUGCAUACC-CAA--CAGGCGCCAGGC---- -..................(((...............---------(((((..(((((...)))))..)))))....(-(..--..))....))).---- ( -12.30) >DroYak_CAF1 144516 89 - 1 UCUCCCACGAGCU--------CCUCUUCCCUUACCAAUUUCCUAUCCUGGCUGGGAAAAGGAUGUUUUGUUGCCAAAC-CAA--CAGGCGCUAGACCGCU ........(((((--------(((.(((((...(((...........)))..))))).)))).))))(((((......-)))--))((((......)))) ( -22.40) >consensus UCUUCCACGACCA________CCUCUUCCCUUACCAAUUUACUAUCCUGGCUGGGAAAAGGAUGUUUUGUUGCCCAAC_CAA__CAGGCGCUAGACCGCU .....................(((.(((((...(((...........)))..))))).)))....((((.((((............)))).))))..... (-11.24 = -12.18 + 0.95)

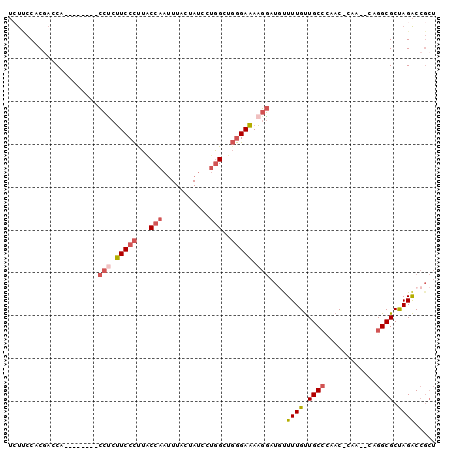
| Location | 16,690,066 – 16,690,161 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 74.27 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -17.27 |
| Energy contribution | -17.15 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16690066 95 + 22407834 GCAACAAAACAUCCUUUUCCCAGCCACGAUGGGAAAUUGGUAAGGGAAGGGGUGGGGUGGUCGUGGAAUAGGCGGAGGGACAGA--GGGGAGCCGAG ....((...(((((((((((..((((...........))))..)))))))))))...))(((........)))...........--((....))... ( -25.80) >DroSim_CAF1 144726 83 + 1 GCAACAAAACAUCCUUUUCCCAGCCAGGAUAUUAAAUUGGUAAGGGAAGAGG-----UGGUAGUGGAAGAUGGAGAG---------GGGGGGCCGAG ....((..((..(((((((((.(((((.........)))))..)))))))))-----..))..))...........(---------(.....))... ( -21.70) >DroEre_CAF1 145229 79 + 1 GCAACAAAGCAUCCUUUUCCCAGCCAGGAUGGCAAGUUGGCAGGGGAAGAGG-----AGCUCGUGGGAGAG-------------AGUGGGUGGCGAG ((.((...((.((((((((((.(((((.........)))))..)))))))))-----).(((......)))-------------.))..)).))... ( -29.70) >DroYak_CAF1 144540 85 + 1 GCAACAAAACAUCCUUUUCCCAGCCAGGAUAGGAAAUUGGUAAGGGAAGAGG-----AGCUCGUGGGAGAGCG------UUAG-AGGGGGAGUCGAG .......(((.((((((((((.(((((.........)))))..)))))))))-----)((((......)))))------))..-............. ( -27.60) >consensus GCAACAAAACAUCCUUUUCCCAGCCAGGAUAGGAAAUUGGUAAGGGAAGAGG_____AGCUCGUGGAAGAGGG_____________GGGGAGCCGAG ............(((((((((.(((((.........)))))..)))))))))............................................. (-17.27 = -17.15 + -0.12)

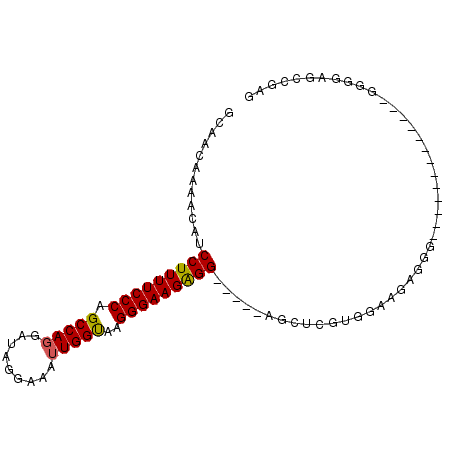
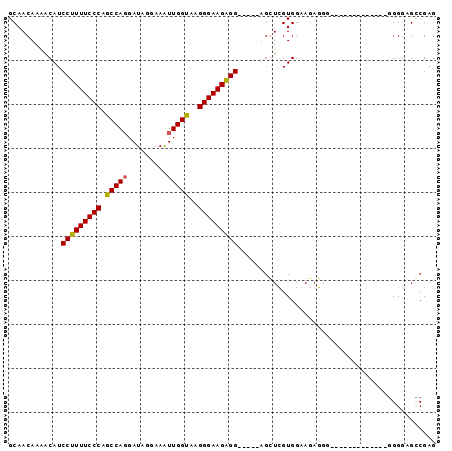
| Location | 16,690,066 – 16,690,161 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 74.27 |
| Mean single sequence MFE | -17.87 |
| Consensus MFE | -11.52 |
| Energy contribution | -11.78 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16690066 95 - 22407834 CUCGGCUCCCC--UCUGUCCCUCCGCCUAUUCCACGACCACCCCACCCCUUCCCUUACCAAUUUCCCAUCGUGGCUGGGAAAAGGAUGUUUUGUUGC ...(((.....--...........))).......((((.....((((..(((((...(((...........)))..)))))..)).))....)))). ( -14.89) >DroSim_CAF1 144726 83 - 1 CUCGGCCCCCC---------CUCUCCAUCUUCCACUACCA-----CCUCUUCCCUUACCAAUUUAAUAUCCUGGCUGGGAAAAGGAUGUUUUGUUGC ...........---------....................-----(((.(((((...(((...........)))..))))).)))............ ( -12.90) >DroEre_CAF1 145229 79 - 1 CUCGCCACCCACU-------------CUCUCCCACGAGCU-----CCUCUUCCCCUGCCAACUUGCCAUCCUGGCUGGGAAAAGGAUGCUUUGUUGC .............-------------.........(((((-----(((.(((((..((((...........)))).))))).)))).))))...... ( -22.40) >DroYak_CAF1 144540 85 - 1 CUCGACUCCCCCU-CUAA------CGCUCUCCCACGAGCU-----CCUCUUCCCUUACCAAUUUCCUAUCCUGGCUGGGAAAAGGAUGUUUUGUUGC ..((((.......-....------.((((......))))(-----(((.(((((...(((...........)))..))))).))))......)))). ( -21.30) >consensus CUCGGCUCCCC_____________CCCUCUCCCACGACCA_____CCUCUUCCCUUACCAAUUUCCCAUCCUGGCUGGGAAAAGGAUGUUUUGUUGC .............................................(((.(((((...(((...........)))..))))).)))............ (-11.52 = -11.78 + 0.25)

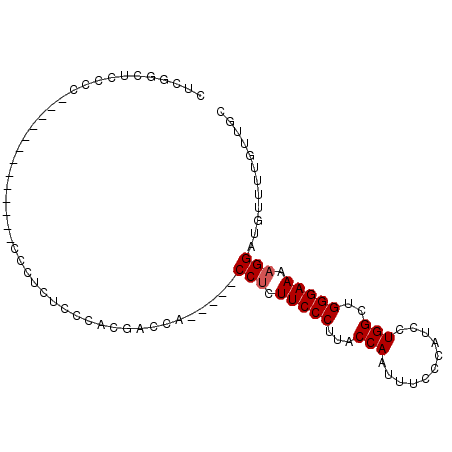
| Location | 16,690,136 – 16,690,226 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 79.46 |
| Mean single sequence MFE | -14.58 |
| Consensus MFE | -14.01 |
| Energy contribution | -13.57 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
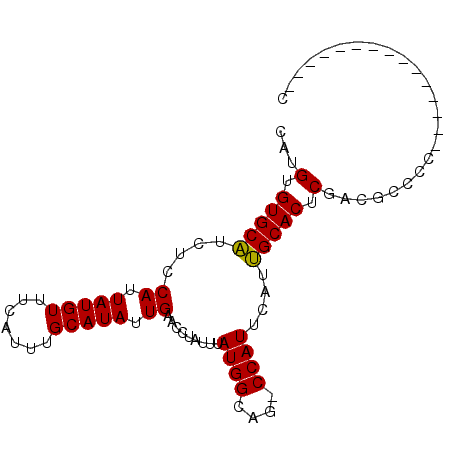
>2L_DroMel_CAF1 16690136 90 - 22407834 CAUGUGUGCAUCUCCAUUAUGUUUCAUUUGCAUAUUGAACCUAUUUAUGGCAG-CCAUUCAUUGCACUCGGCUCCCC--UCUGUCCCUCCGCC ...(.(((((....((.(((((.......))))).)).........((((...-))))....))))).)(((.....--...)))........ ( -13.80) >DroPse_CAF1 161195 75 - 1 UGUGUGUGCCUUUCCAUUAUGUUCCAUUUGCAUAUUGAACCUAUUUAUGGCAACCCAUUCAUGGCACUCCAUGCC------------------ .(((.(((((....((.(((((.......))))).)).........((((....))))....)))))..)))...------------------ ( -15.40) >DroSim_CAF1 144791 83 - 1 CAUGUGUGCAUCUCCAUUAUGUUUCAUUUGCAUAUUGAACCUAUUUAUGGCAG-CCAUUCAUUGCACUCGGCCCCCC---------CUCUCCA ..((.(((((....((.(((((.......))))).)).........((((...-))))....))))).)).......---------....... ( -12.60) >DroEre_CAF1 145294 79 - 1 CAAGUGUGCAUCUCCAUUAUGUUGCAUUUGCAUAUUGAACCUAUUUAUGGCAG-CCAUUCAUUGCACUCGCCACCCACU-------------C ..((((((((....((......))....))))))))...........((((((-.((.....))..)).))))......-------------. ( -12.90) >DroYak_CAF1 144605 85 - 1 CAUGUGUGCAUUUCCAUUAUGUUUCAUUUGCAUAUUGGACCUAUUUAUGGCAG-CCAUUCAUUGCACUCGACUCCCCCU-CUAA------CGC ..((.(((((..((((.(((((.......))))).)))).......((((...-))))....))))).)).........-....------... ( -17.10) >DroPer_CAF1 163098 75 - 1 UAUGUGUGCCUUUCCAUUAUGUUCCAUUUGCAUAUUGAACCUAUUUAUGGCAACCCAUUCAUGGCACUCCAUGCC------------------ ((((.(((((....((.(((((.......))))).)).........((((....))))....)))))..))))..------------------ ( -15.70) >consensus CAUGUGUGCAUCUCCAUUAUGUUUCAUUUGCAUAUUGAACCUAUUUAUGGCAG_CCAUUCAUUGCACUCGACGCCCC_______________C ...(.(((((....((.(((((.......))))).)).........((((....))))....))))).)........................ (-14.01 = -13.57 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:05 2006