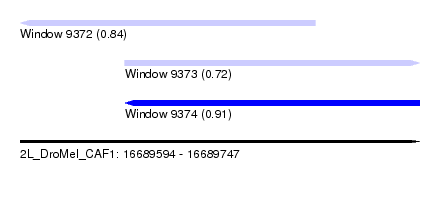
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,689,594 – 16,689,747 |
| Length | 153 |
| Max. P | 0.911479 |

| Location | 16,689,594 – 16,689,707 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.88 |
| Mean single sequence MFE | -23.48 |
| Consensus MFE | -7.36 |
| Energy contribution | -7.08 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16689594 113 - 22407834 AUAUAUUUGAUUUACAGAAAAAUAUCUGACAGCCCAAUUAUCAGUAAGGGAGUCAAAUUUUAUUUACUGAA-------AAUUUAUCCGACAGGAUGUUAAUUGGUGUUAAGUGGAAUAAA ...(((((.((((.((((......))))..((((((((((((((((((.(((......))).)))))))).-------....(((((....))))).))))))).))))))).))))).. ( -28.60) >DroSec_CAF1 144351 119 - 1 AUAUAUUUGAUUUACAGAAAAAUAUCUGACAGC-CAAUUAUCAGUAAGGGAGUCAAAUUUUAUUUACUGAUAUUACUGAAUUUAUCUGACAGGAUGUUAAUUGGUGUUAAGUGGAAUAAA ...(((((.(((((((((......))))...((-((((((((((((((.(((......))).))))))))............(((((....))))).))))))))..))))).))))).. ( -26.80) >DroSim_CAF1 144286 119 - 1 AUAUAUUUGAUUUACAGAAAAAUAUCUGACAGC-CAAUUAUCAGUAAGGGAGUCAAAUUUUAUUUACUGAUAUAACUGAAUUUAUCUGACAGGAUGUUAAUUGGUGUUAAGUGGAAUAAA ...(((((.(((((((((......))))...((-((((((((((((((.(((......))).))))))))............(((((....))))).))))))))..))))).))))).. ( -26.80) >DroEre_CAF1 144814 100 - 1 AUAAAUUCGAUGUACAAUAAAAUAUCUGACAGU-CUAUAAU------------CAGAUAUUAUGUACUGCAAU-------UUUAUCUGACAGGAUGAUAAUUAGUGUUGAAUUGAAUAAA ...(((((((((((((....(((((((((....-......)------------)))))))).))))).(((((-------(((((((....)))))).)))).))))))))))....... ( -21.30) >DroYak_CAF1 144097 98 - 1 AUAAAUUCGAUGUACAGUAAAAUAUCUGUCAGC-CUUU-AU------------CAUAUAUUAUUUCCUGCUAU-------UUUAUCUGACAGGAUGUUAAUUGGUGUUAAAUAGAA-AAA .....(((..((.((((........))))))..-.(((-(.------------(((..((((..((((((...-------.......).)))))...))))..))).))))..)))-... ( -13.90) >consensus AUAUAUUUGAUUUACAGAAAAAUAUCUGACAGC_CAAUUAUCAGUAAGGGAGUCAAAUUUUAUUUACUGAUAU_____AAUUUAUCUGACAGGAUGUUAAUUGGUGUUAAGUGGAAUAAA ...((((((((...(((........)))......................................................(((((....))))).........))))))))....... ( -7.36 = -7.08 + -0.28)

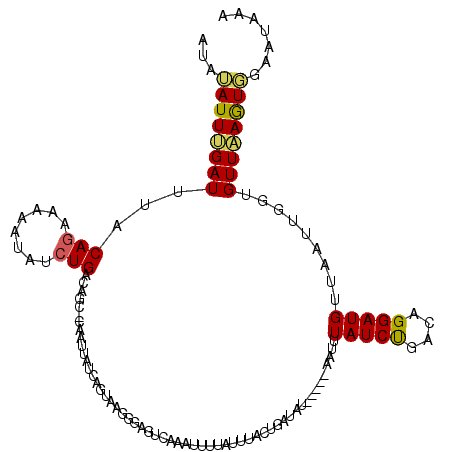
| Location | 16,689,634 – 16,689,747 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.68 |
| Mean single sequence MFE | -24.46 |
| Consensus MFE | -8.12 |
| Energy contribution | -8.80 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16689634 113 + 22407834 UU-------UUCAGUAAAUAAAAUUUGACUCCCUUACUGAUAAUUGGGCUGUCAGAUAUUUUUCUGUAAAUCAAAUAUAUUGUGAUGACAGCUGAAUGAGCAUGUUAAAUGCAGUCAAUU ..-------...............((((((............(((.((((((((((......)))....((((.........))))))))))).)))..((((.....)))))))))).. ( -24.60) >DroSec_CAF1 144391 119 + 1 UUCAGUAAUAUCAGUAAAUAAAAUUUGACUCCCUUACUGAUAAUUG-GCUGUCAGAUAUUUUUCUGUAAAUCAAAUAUAUUGUGAUGACAGCUGAAUGAGCAAGUUAAAUGCAGUCAAUU .(((....(((((((((................))))))))).(..-(((((((((......)))....((((.........))))))))))..).)))(((.......)))........ ( -24.39) >DroSim_CAF1 144326 119 + 1 UUCAGUUAUAUCAGUAAAUAAAAUUUGACUCCCUUACUGAUAAUUG-GCUGUCAGAUAUUUUUCUGUAAAUCAAAUAUAUUGUGAUGACAGCUGAAUGAGCAAGUUAAAUGCAGUCAAUU .(((....(((((((((................))))))))).(..-(((((((((......)))....((((.........))))))))))..).)))(((.......)))........ ( -24.39) >DroEre_CAF1 144854 100 + 1 -------AUUGCAGUACAUAAUAUCUG------------AUUAUAG-ACUGUCAGAUAUUUUAUUGUACAUCGAAUUUAUUGUGAUGACAGCUGAAUGUGUCAGUUAAAUUCAGUUGAUU -------..(((((((...((((((((------------((.....-...)))))))))).)))))))(((((.........))))).(((((((((...........)))))))))... ( -28.40) >DroYak_CAF1 144136 99 + 1 -------AUAGCAGGAAAUAAUAUAUG------------AU-AAAG-GCUGACAGAUAUUUUACUGUACAUCGAAUUUAUUGUGAUGACAGCUGAAUGGGUCAGUUAUGGGCAGUCAAUU -------..................((------------..-.(..-((((((...(((((..((((.(((((.........)))))))))..))))).))))))..)...))....... ( -20.50) >consensus UU_____AUAUCAGUAAAUAAAAUUUGACUCCCUUACUGAUAAUUG_GCUGUCAGAUAUUUUUCUGUAAAUCAAAUAUAUUGUGAUGACAGCUGAAUGAGCAAGUUAAAUGCAGUCAAUU .........................((((..................(((((((.(((......((.....)).......)))..))))))).......(((.......))).))))... ( -8.12 = -8.80 + 0.68)

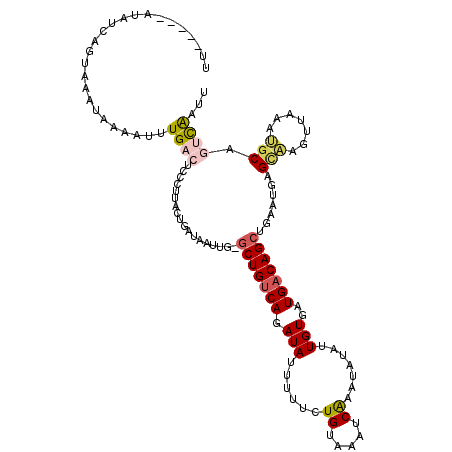
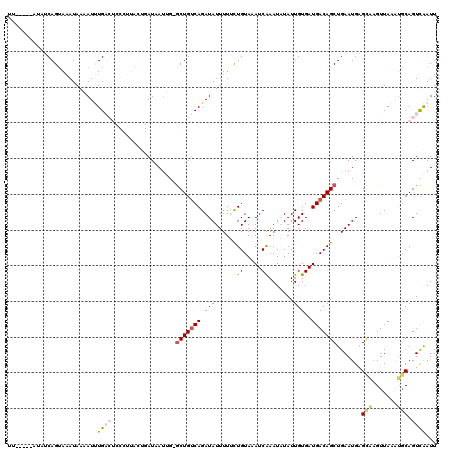
| Location | 16,689,634 – 16,689,747 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.68 |
| Mean single sequence MFE | -20.42 |
| Consensus MFE | -6.02 |
| Energy contribution | -6.06 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.29 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
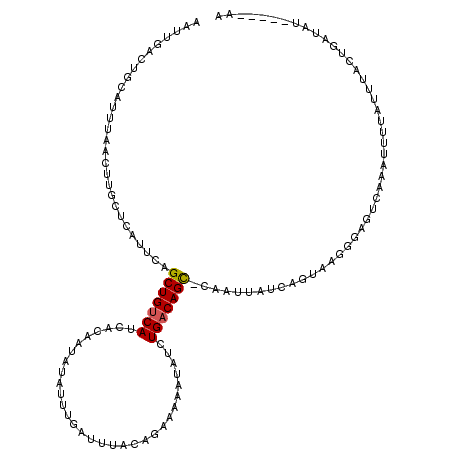
>2L_DroMel_CAF1 16689634 113 - 22407834 AAUUGACUGCAUUUAACAUGCUCAUUCAGCUGUCAUCACAAUAUAUUUGAUUUACAGAAAAAUAUCUGACAGCCCAAUUAUCAGUAAGGGAGUCAAAUUUUAUUUACUGAA-------AA (((((...((((.....)))).......((((((....(((.....)))......(((......))))))))).))))).((((((((.(((......))).)))))))).-------.. ( -22.70) >DroSec_CAF1 144391 119 - 1 AAUUGACUGCAUUUAACUUGCUCAUUCAGCUGUCAUCACAAUAUAUUUGAUUUACAGAAAAAUAUCUGACAGC-CAAUUAUCAGUAAGGGAGUCAAAUUUUAUUUACUGAUAUUACUGAA ...(((..(((.......))))))((((((((((....(((.....)))......(((......)))))))))-....((((((((((.(((......))).))))))))))....)))) ( -23.70) >DroSim_CAF1 144326 119 - 1 AAUUGACUGCAUUUAACUUGCUCAUUCAGCUGUCAUCACAAUAUAUUUGAUUUACAGAAAAAUAUCUGACAGC-CAAUUAUCAGUAAGGGAGUCAAAUUUUAUUUACUGAUAUAACUGAA ...(((..(((.......))))))((((((((((....(((.....)))......(((......)))))))))-....((((((((((.(((......))).))))))))))....)))) ( -23.70) >DroEre_CAF1 144854 100 - 1 AAUCAACUGAAUUUAACUGACACAUUCAGCUGUCAUCACAAUAAAUUCGAUGUACAAUAAAAUAUCUGACAGU-CUAUAAU------------CAGAUAUUAUGUACUGCAAU------- .......((((((((..(((((........)))))......))))))))..(((((....(((((((((....-......)------------)))))))).)))))......------- ( -18.30) >DroYak_CAF1 144136 99 - 1 AAUUGACUGCCCAUAACUGACCCAUUCAGCUGUCAUCACAAUAAAUUCGAUGUACAGUAAAAUAUCUGUCAGC-CUUU-AU------------CAUAUAUUAUUUCCUGCUAU------- ................(((((..(((..(((((((((...........)))).)))))..)))....))))).-....-..------------....................------- ( -13.70) >consensus AAUUGACUGCAUUUAACUUGCUCAUUCAGCUGUCAUCACAAUAUAUUUGAUUUACAGAAAAAUAUCUGACAGC_CAAUUAUCAGUAAGGGAGUCAAAUUUUAUUUACUGAUAU_____AA ............................(((((((...............................)))))))............................................... ( -6.02 = -6.06 + 0.04)

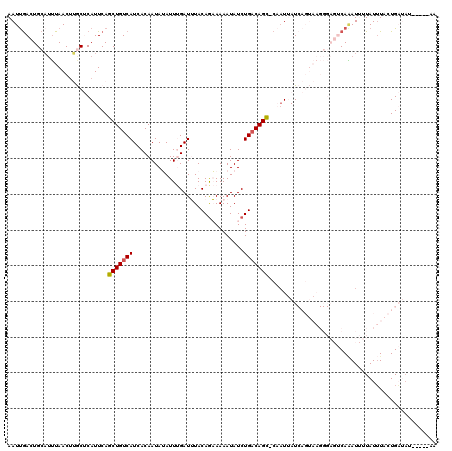
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:01 2006