
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,685,056 – 16,685,238 |
| Length | 182 |
| Max. P | 0.958139 |

| Location | 16,685,056 – 16,685,169 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 83.28 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -15.16 |
| Energy contribution | -15.72 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16685056 113 + 22407834 AUUUCCCUUUGCCUACGUUUCCUCCACUCUUUCCAUUAGGCUGGCCAUCACAUUUGAGGUUUUAUCCUUGGGCUGUCAACACGUGCUU--UGACACGCCCCCUUUAGAAUCCUCC .....((...(((((.....................))))).))...........(((((((((.....((((((((((........)--))))).))))....))))).)))). ( -26.50) >DroSec_CAF1 139915 113 + 1 AUUUCCCUUUGCCUACGUUUCCUCCUCUCUUCCCAUUAGACUGGCCAUCACUUUUGAGGUUUUAUCCUUGGGCUGUCAACACGUGAUU--UGACACGCCCCCUUUAGAAUCCUCC ..........(((...((((.................)))).)))..........(((((((((.....((((((((((........)--))))).))))....))))).)))). ( -23.33) >DroSim_CAF1 139838 113 + 1 AUUUCCCUUUGCCUACGUUUCCUCCUCUCUUCCCAUUAGGCUGGCCAUCACUUUUGAGGUUUUAUCCUUGGGCUGUCAACACGUGCUU--UGACACGCCCCCUUUAGAAUCCUCC .....((...(((((.....................))))).))...........(((((((((.....((((((((((........)--))))).))))....))))).)))). ( -26.50) >DroEre_CAF1 140457 113 + 1 AUUUCCCAUCGCCUACGUUUCCUCCUCUCUUCCCAUUAGGCUGCCCAUCACUUUUGAGGUUUUAUCCUUGGGCUGUCAACACGUGCUU--UGACACGCCCCCUUUAGCGUCCUCC ..........(((((.....................)))))..............((((..........((((((((((........)--))))).))))..........)))). ( -23.15) >DroYak_CAF1 139720 113 + 1 AUUUUCCUUUGCCUACGUUUCCUCCUCUCCUUCCAUUAGGCUGCCCAUCACUUUUGAGGUUUUAUCCUUGGGCUGUCAACACGUGCUU--UGACACGCCCCUUUAAGCGUCCUUC ..........(((((.....................))))).((...........((((......))))((((((((((........)--))))).))))......))....... ( -22.30) >DroPer_CAF1 157032 91 + 1 AC-UCCCCCA------------GCCUCCCUUUUCAUU-------CCAUCACUUUUGAGGUUUUGGCUUUAGGCUGUCGACACGUGCUGUGUGACACGCCCCGUU----GUCCCCC ..-......(------------(((..((((......-------...........))))....))))...(((((((.((((.....)))))))).))).....----....... ( -19.83) >consensus AUUUCCCUUUGCCUACGUUUCCUCCUCUCUUCCCAUUAGGCUGGCCAUCACUUUUGAGGUUUUAUCCUUGGGCUGUCAACACGUGCUU__UGACACGCCCCCUUUAGAAUCCUCC ..........(((((.....................)))))..............((((..........(((((((((............))))).))))..........)))). (-15.16 = -15.72 + 0.56)

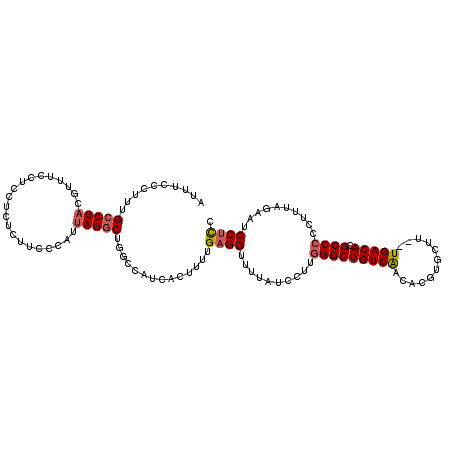
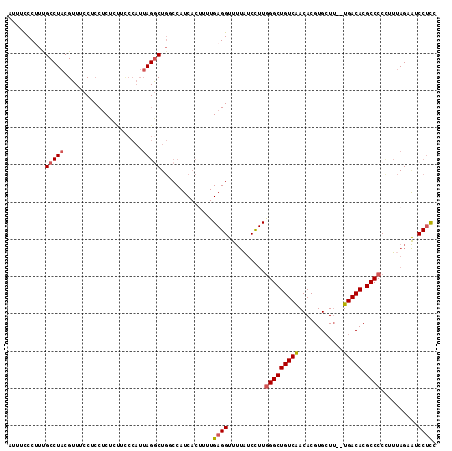
| Location | 16,685,056 – 16,685,169 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 83.28 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -19.44 |
| Energy contribution | -19.93 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16685056 113 - 22407834 GGAGGAUUCUAAAGGGGGCGUGUCA--AAGCACGUGUUGACAGCCCAAGGAUAAAACCUCAAAUGUGAUGGCCAGCCUAAUGGAAAGAGUGGAGGAAACGUAGGCAAAGGGAAAU .(((((((((.....((((.(((((--(........)))))))))).)))))....))))..........(((..((....))..........(....)...))).......... ( -28.90) >DroSec_CAF1 139915 113 - 1 GGAGGAUUCUAAAGGGGGCGUGUCA--AAUCACGUGUUGACAGCCCAAGGAUAAAACCUCAAAAGUGAUGGCCAGUCUAAUGGGAAGAGAGGAGGAAACGUAGGCAAAGGGAAAU .(((((((((.....((((.(((((--(........)))))))))).)))))....))))..........(((..(((....)))........(....)...))).......... ( -27.90) >DroSim_CAF1 139838 113 - 1 GGAGGAUUCUAAAGGGGGCGUGUCA--AAGCACGUGUUGACAGCCCAAGGAUAAAACCUCAAAAGUGAUGGCCAGCCUAAUGGGAAGAGAGGAGGAAACGUAGGCAAAGGGAAAU .(((((((((.....((((.(((((--(........)))))))))).)))))....))))..........(((..((....))..........(....)...))).......... ( -29.10) >DroEre_CAF1 140457 113 - 1 GGAGGACGCUAAAGGGGGCGUGUCA--AAGCACGUGUUGACAGCCCAAGGAUAAAACCUCAAAAGUGAUGGGCAGCCUAAUGGGAAGAGAGGAGGAAACGUAGGCGAUGGGAAAU ......((((.....((((.(((((--(........))))))(((((.((......))(((....)))))))).))))...............(....)...))))......... ( -28.50) >DroYak_CAF1 139720 113 - 1 GAAGGACGCUUAAAGGGGCGUGUCA--AAGCACGUGUUGACAGCCCAAGGAUAAAACCUCAAAAGUGAUGGGCAGCCUAAUGGAAGGAGAGGAGGAAACGUAGGCAAAGGAAAAU .......(((((...((((.(((((--(........))))))))))..........((((....((.....))..(((......))).)))).(....).))))).......... ( -28.60) >DroPer_CAF1 157032 91 - 1 GGGGGAC----AACGGGGCGUGUCACACAGCACGUGUCGACAGCCUAAAGCCAAAACCUCAAAAGUGAUGG-------AAUGAAAAGGGAGGC------------UGGGGGA-GU .(....)----..(((.((((((......)))))).))).((((((....((....(((((....))).))-------..(....))).))))------------)).....-.. ( -26.70) >consensus GGAGGACUCUAAAGGGGGCGUGUCA__AAGCACGUGUUGACAGCCCAAGGAUAAAACCUCAAAAGUGAUGGCCAGCCUAAUGGAAAGAGAGGAGGAAACGUAGGCAAAGGGAAAU .((((..........((((.(((((....((....)))))))))))..........))))..............(((((..............(....).))))).......... (-19.44 = -19.93 + 0.49)

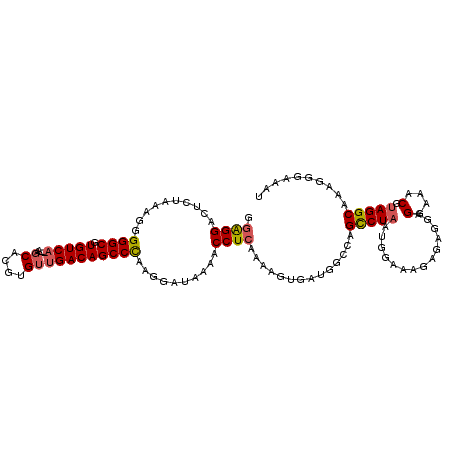

| Location | 16,685,096 – 16,685,203 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -25.99 |
| Consensus MFE | -17.69 |
| Energy contribution | -17.17 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16685096 107 + 22407834 CUGGCCAUCACAUUUGAGGUUU----UAUCCUUGGGCUGUCAACACGUGCUUUGACACGCCCCCUUUAGAAUCCUCCGGGUAUUUUUUGUACCAGUUUUCAGUUAGGAAGU (((((..........(((((((----((.....((((((((((........)))))).))))....))))).))))..(((((.....)))))........)))))..... ( -29.70) >DroSec_CAF1 139955 107 + 1 CUGGCCAUCACUUUUGAGGUUU----UAUCCUUGGGCUGUCAACACGUGAUUUGACACGCCCCCUUUAGAAUCCUCCGGGUAUUUUUUGUACCACUUUUCAGUUAGGAUGU (((((..........(((((((----((.....((((((((((........)))))).))))....))))).))))..(((((.....)))))........)))))..... ( -29.70) >DroSim_CAF1 139878 107 + 1 CUGGCCAUCACUUUUGAGGUUU----UAUCCUUGGGCUGUCAACACGUGCUUUGACACGCCCCCUUUAGAAUCCUCCGGGUAUUUUUUGUACCACUUUUCAGUUAGGAUGU (((((..........(((((((----((.....((((((((((........)))))).))))....))))).))))..(((((.....)))))........)))))..... ( -29.70) >DroEre_CAF1 140497 107 + 1 CUGCCCAUCACUUUUGAGGUUU----UAUCCUUGGGCUGUCAACACGUGCUUUGACACGCCCCCUUUAGCGUCCUCCCGGUGUUUGUUGUACCACUUUUCAGUUAGGAAGU .(((.((.((((...((((...----.......((((((((((........)))))).))))..........))))..))))..))..)))..((((((......)))))) ( -24.35) >DroYak_CAF1 139760 103 + 1 CUGCCCAUCACUUUUGAGGUUU----UAUCCUUGGGCUGUCAACACGUGCUUUGACACGCCCCUUUAAGCGUCCUUCUGGUAUUU-UUGUACUACUUUUUAGUC---AAAU .((((..........((((...----...))))((((((((((........)))))).))))................))))..(-(((.((((.....)))))---))). ( -22.80) >DroAna_CAF1 79756 94 + 1 -----------UUUUGAGGUUUUUUUUGUCUCUCGGCUGUCGACACGUGCUUUGACACGCCCCCUUUUGGACAGCCCGC-CCCCCGUUGGAGGAAGUUUAA-----AUAGU -----------.((((((.(((((((........(((((((....((((......))))....(....))))))))...-........))))))).)))))-----).... ( -19.71) >consensus CUGGCCAUCACUUUUGAGGUUU____UAUCCUUGGGCUGUCAACACGUGCUUUGACACGCCCCCUUUAGAAUCCUCCGGGUAUUUUUUGUACCACUUUUCAGUUAGGAAGU ...............((((..........))))((((((((((........)))))).))))................(((((.....))))).................. (-17.69 = -17.17 + -0.53)

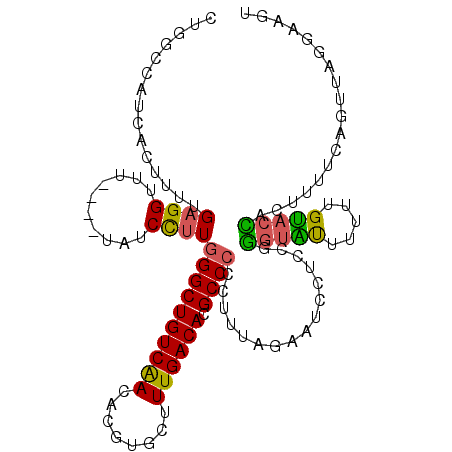
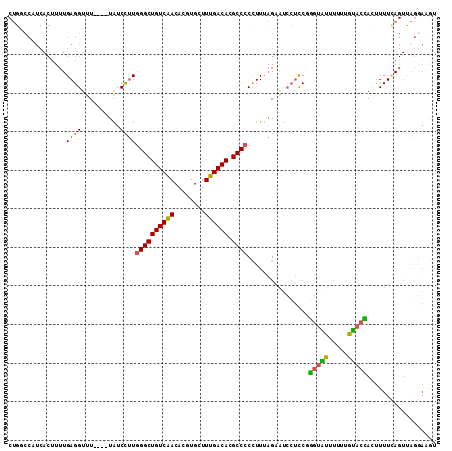
| Location | 16,685,096 – 16,685,203 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -28.08 |
| Consensus MFE | -14.77 |
| Energy contribution | -15.38 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.642096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16685096 107 - 22407834 ACUUCCUAACUGAAAACUGGUACAAAAAAUACCCGGAGGAUUCUAAAGGGGGCGUGUCAAAGCACGUGUUGACAGCCCAAGGAUA----AAACCUCAAAUGUGAUGGCCAG ................(((((.......(((....(((((((((.....((((.((((((........)))))))))).))))).----...))))...)))....))))) ( -26.10) >DroSec_CAF1 139955 107 - 1 ACAUCCUAACUGAAAAGUGGUACAAAAAAUACCCGGAGGAUUCUAAAGGGGGCGUGUCAAAUCACGUGUUGACAGCCCAAGGAUA----AAACCUCAAAAGUGAUGGCCAG .((((((.(((....)))((((.......))))..(((((((((.....((((.((((((........)))))))))).))))).----...))))...)).))))..... ( -28.90) >DroSim_CAF1 139878 107 - 1 ACAUCCUAACUGAAAAGUGGUACAAAAAAUACCCGGAGGAUUCUAAAGGGGGCGUGUCAAAGCACGUGUUGACAGCCCAAGGAUA----AAACCUCAAAAGUGAUGGCCAG .((((((.(((....)))((((.......))))..(((((((((.....((((.((((((........)))))))))).))))).----...))))...)).))))..... ( -28.90) >DroEre_CAF1 140497 107 - 1 ACUUCCUAACUGAAAAGUGGUACAACAAACACCGGGAGGACGCUAAAGGGGGCGUGUCAAAGCACGUGUUGACAGCCCAAGGAUA----AAACCUCAAAAGUGAUGGGCAG .((((((.(((....)))(((.........)))))))))(((((......)))))(((((........))))).(((((.((...----...))(((....)))))))).. ( -30.90) >DroYak_CAF1 139760 103 - 1 AUUU---GACUAAAAAGUAGUACAA-AAAUACCAGAAGGACGCUUAAAGGGGCGUGUCAAAGCACGUGUUGACAGCCCAAGGAUA----AAACCUCAAAAGUGAUGGGCAG .(((---(((((.....)))).)))-)....((....))..(((((...((((.((((((........)))))))))).(((...----...))).........))))).. ( -24.60) >DroAna_CAF1 79756 94 - 1 ACUAU-----UUAAACUUCCUCCAACGGGGG-GCGGGCUGUCCAAAAGGGGGCGUGUCAAAGCACGUGUCGACAGCCGAGAGACAAAAAAAACCUCAAAA----------- .....-----.....(.((((((...)))))-).)((((((((....)((.((((((....)))))).)))))))))..(((...........)))....----------- ( -29.10) >consensus ACUUCCUAACUGAAAAGUGGUACAAAAAAUACCCGGAGGAUUCUAAAGGGGGCGUGUCAAAGCACGUGUUGACAGCCCAAGGAUA____AAACCUCAAAAGUGAUGGCCAG ..................((((.......))))..((((...(.....)((((.((((((........))))))))))..............))))............... (-14.77 = -15.38 + 0.61)



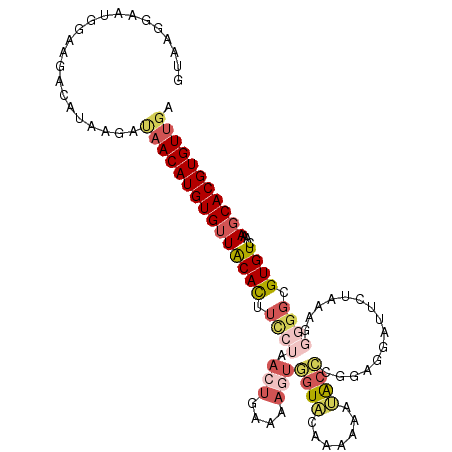
| Location | 16,685,131 – 16,685,238 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 75.98 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -16.32 |
| Energy contribution | -17.85 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16685131 107 - 22407834 GUGAGGAAUGGAAGACAUAAGACAACAUGUGUUACACUUCCUAACUGAAAACUGGUACAAAAAAUACCCGGAGGAUUCUAAAGGGGGCGUGUCAAAGCACGUGUUGA .......(((.....)))....(((((((((((((((.((((....(((..((((((.......))))...))..))).....)))).))))...))))))))))). ( -28.50) >DroSec_CAF1 139990 107 - 1 GUGAGGAAUGGAAGACAUAAGACAACAUGUGUUACACAUCCUAACUGAAAAGUGGUACAAAAAAUACCCGGAGGAUUCUAAAGGGGGCGUGUCAAAUCACGUGUUGA .......(((.....)))....(((((((((..((((..(((.(((....)))((((.......))))...............)))..)))).....))))))))). ( -25.20) >DroSim_CAF1 139913 100 - 1 GU-------GGAAGACAUAAAAGAACAUGUGUUACACAUCCUAACUGAAAAGUGGUACAAAAAAUACCCGGAGGAUUCUAAAGGGGGCGUGUCAAAGCACGUGUUGA ((-------(.....))).....((((((((((((((..(((.(((....)))((((.......))))...............)))..))))...)))))))))).. ( -24.70) >DroEre_CAF1 140532 107 - 1 GUAAGGAAUGGAAGACAGAAUUUAACAUGUGUUACACUUCCUAACUGAAAAGUGGUACAACAAACACCGGGAGGACGCUAAAGGGGGCGUGUCAAAGCACGUGUUGA .....................((((((((((((...((((((.(((....)))(((.........)))))))))(((((......))))).....)))))))))))) ( -30.40) >DroYak_CAF1 139795 103 - 1 GUACGAAAGGAAAGACAUGAUUUAACAUGUGUUACAUUU---GACUAAAAAGUAGUACAA-AAAUACCAGAAGGACGCUUAAAGGGGCGUGUCAAAGCACGUGUUGA ...(....)............(((((((((((....(((---(((...............-.....((....))((((((....))))))))))))))))))))))) ( -25.70) >DroAna_CAF1 79784 99 - 1 UCCGAGCCUG--AGAAACAGGAUAACAUGUGUUGCACUAU-----UUAAACUUCCUCCAACGGGGG-GCGGGCUGUCCAAAAGGGGGCGUGUCAAAGCACGUGUCGA ..(((.((((--.....))))....((((((((((((...-----.....((.(((((....))))-).))...((((......))))))))...))))))))))). ( -33.10) >consensus GUAAGGAAUGGAAGACAUAAGAUAACAUGUGUUACACUUCCUAACUGAAAAGUGGUACAAAAAAUACCCGGAGGAUUCUAAAGGGGGCGUGUCAAAGCACGUGUUGA ......................(((((((((((((((.((((.(((....)))((((.......))))...............)))).))))...))))))))))). (-16.32 = -17.85 + 1.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:58 2006