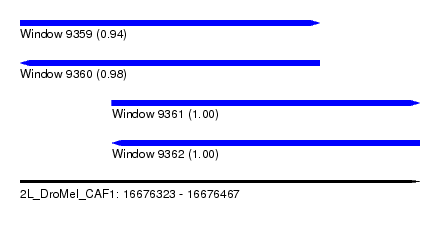
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,676,323 – 16,676,467 |
| Length | 144 |
| Max. P | 0.998652 |

| Location | 16,676,323 – 16,676,431 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.87 |
| Mean single sequence MFE | -26.35 |
| Consensus MFE | -11.84 |
| Energy contribution | -13.07 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16676323 108 + 22407834 GUUUAACUUUUUGGCGCAAUAUUAUUCACCUUGACCACGCACCGGUGUGCAAAAGAUUCACCGCA---CACCGGUGCACUUAAAUUCACCACAGCCUUAAUAAUUCUUACG ............(((..........((.....))....((((((((((((............)))---)))))))))................)))............... ( -29.50) >DroSec_CAF1 131062 108 + 1 GUUUAACUUUUUGGCGCAAUAUUAUUCACCUUGACCACGCACCGGUGUGCAAAAGAUUAACCGCA---CAUCGGUGCAUUUAAAUUCACCACAGCCGUAAUAAUUCCUACG ............(((..........((.....))....((((((((((((............)))---)))))))))................)))(((........))). ( -27.80) >DroSim_CAF1 122918 108 + 1 GUUUAACUUUUUGGCGCAAUAUUAUUCACCUUGACCACGCACCGGUGUGCAAAAGAUUCACCGCA---CAUCGGUGCAUUUAAAUUCACCACAGCCCUAAUAAUUCCUACG ((........((((.((........((.....))....((((((((((((............)))---)))))))))................)).))))........)). ( -26.79) >DroEre_CAF1 131403 108 + 1 GUUUGACUUUUUGGCGCAAUAAUAUUCACCUUGACCAUACACCGGUGUGCAAAAAAUUCACCGCA---CGCCGGUGCAUACAAAUUCACCACCGCCGUAAUGAUUCUUACG ((..((.((..(((((.........((.....)).....(((((((((((............)))---))))))))................)))))....)).))..)). ( -28.60) >DroYak_CAF1 130401 106 + 1 GUUUGACUUUUUGGCGCAAUAUUAUUCACCUUGACCACACACCGGUGUGCAAA-AAUUCACCGCA---CACCGGUAUAUUGAAAUUCACCACAGUCCUAAUGAUUC-UAUG ....((((...(((.(((((((...((.....))......((((((((((...-........)))---))))))))))))).....).))).))))..........-.... ( -24.30) >DroPer_CAF1 150315 89 + 1 GUUUGACUUUUUGGCGCAAAAUAAUCUACC--------ACACCGGUGAGAGA---ACUCCCCACAGCAUGGUGGCCCAA-----UGCGC-----AAUUU-UUGCCCAAACG .........(((((.((((((.........--------.....((.(((...---.))).))...((((((....)).)-----)))..-----...))-))))))))).. ( -21.10) >consensus GUUUAACUUUUUGGCGCAAUAUUAUUCACCUUGACCACACACCGGUGUGCAAAAGAUUCACCGCA___CACCGGUGCAUUUAAAUUCACCACAGCCCUAAUAAUUCCUACG ............(((..........((.....))....((((((((((((............)))...)))))))))................)))............... (-11.84 = -13.07 + 1.23)

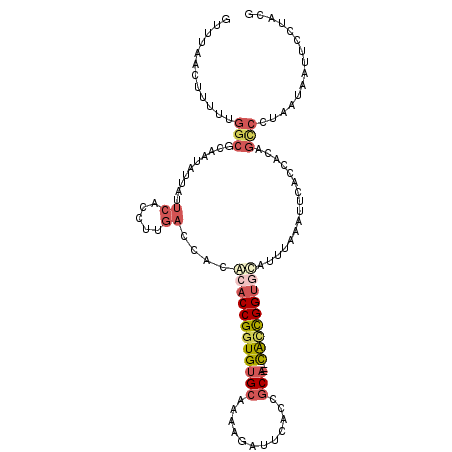
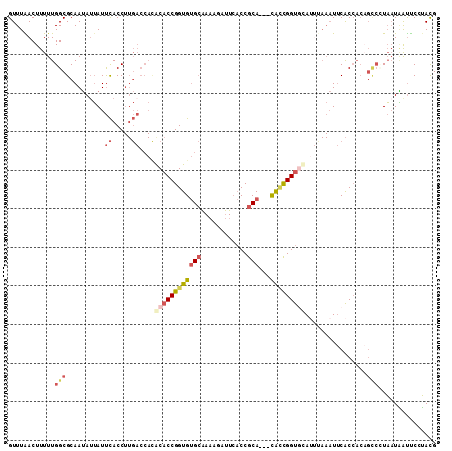
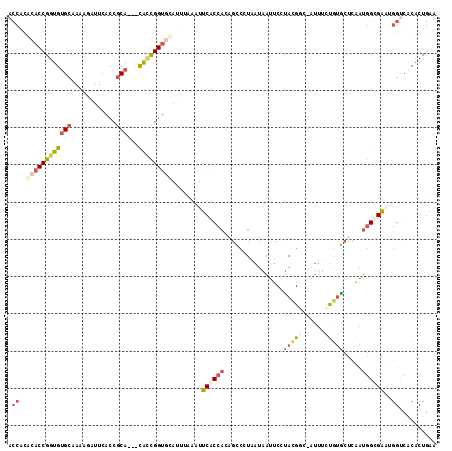
| Location | 16,676,323 – 16,676,431 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.87 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -16.92 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16676323 108 - 22407834 CGUAAGAAUUAUUAAGGCUGUGGUGAAUUUAAGUGCACCGGUG---UGCGGUGAAUCUUUUGCACACCGGUGCGUGGUCAAGGUGAAUAAUAUUGCGCCAAAAAGUUAAAC ...............((((.(((((.........(((((((((---(((((........))))))))))))))....((.....)).........)))))...)))).... ( -36.60) >DroSec_CAF1 131062 108 - 1 CGUAGGAAUUAUUACGGCUGUGGUGAAUUUAAAUGCACCGAUG---UGCGGUUAAUCUUUUGCACACCGGUGCGUGGUCAAGGUGAAUAAUAUUGCGCCAAAAAGUUAAAC (((((......)))))(((.(((((.......((((((((.((---(((((........))))))).))))))))..((.....)).........)))))...)))..... ( -33.90) >DroSim_CAF1 122918 108 - 1 CGUAGGAAUUAUUAGGGCUGUGGUGAAUUUAAAUGCACCGAUG---UGCGGUGAAUCUUUUGCACACCGGUGCGUGGUCAAGGUGAAUAAUAUUGCGCCAAAAAGUUAAAC (((((..((((((...(((....(((......((((((((.((---(((((........))))))).))))))))..))).))).)))))).))))).............. ( -32.10) >DroEre_CAF1 131403 108 - 1 CGUAAGAAUCAUUACGGCGGUGGUGAAUUUGUAUGCACCGGCG---UGCGGUGAAUUUUUUGCACACCGGUGUAUGGUCAAGGUGAAUAUUAUUGCGCCAAAAAGUCAAAC .((((......))))(((.(..((((.....((((((((((.(---(((((........)))))).)))))))))).((.....))...))))..))))............ ( -37.00) >DroYak_CAF1 130401 106 - 1 CAUA-GAAUCAUUAGGACUGUGGUGAAUUUCAAUAUACCGGUG---UGCGGUGAAUU-UUUGCACACCGGUGUGUGGUCAAGGUGAAUAAUAUUGCGCCAAAAAGUCAAAC ....-..........((((.(((((.......(((((((((((---(((((......-.))))))))))))))))..((.....)).........)))))...)))).... ( -37.20) >DroPer_CAF1 150315 89 - 1 CGUUUGGGCAA-AAAUU-----GCGCA-----UUGGGCCACCAUGCUGUGGGGAGU---UCUCUCACCGGUGU--------GGUAGAUUAUUUUGCGCCAAAAAGUCAAAC ..(((((((((-((...-----(.((.-----....)))(((((((((..((((..---..))))..))))))--------)))......)))))).)))))......... ( -28.40) >consensus CGUAAGAAUUAUUAAGGCUGUGGUGAAUUUAAAUGCACCGGUG___UGCGGUGAAUCUUUUGCACACCGGUGCGUGGUCAAGGUGAAUAAUAUUGCGCCAAAAAGUCAAAC ...............((((.((((........((((((((.((...(((((........))))))).)))))))).......((((......))))))))...)))).... (-16.92 = -17.90 + 0.98)

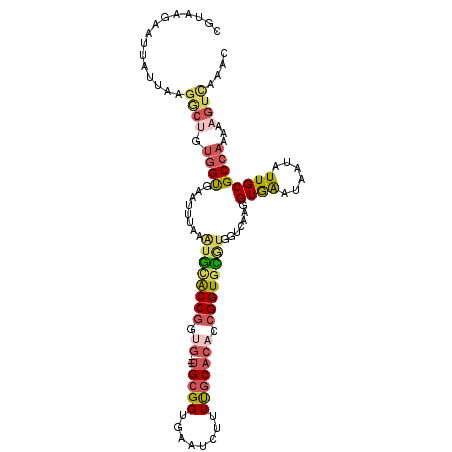
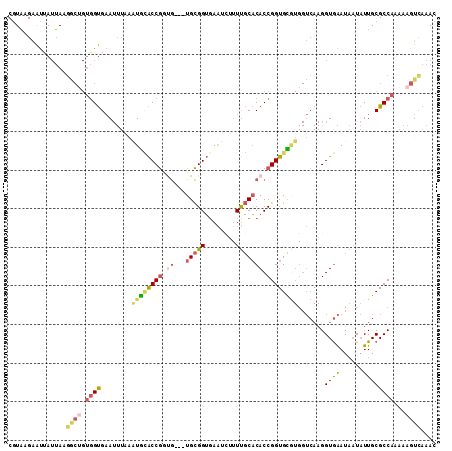
| Location | 16,676,356 – 16,676,467 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 69.30 |
| Mean single sequence MFE | -34.54 |
| Consensus MFE | -14.23 |
| Energy contribution | -15.15 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.48 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.41 |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.998652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16676356 111 + 22407834 ACCACGCACCGGUGUGCAAAAGAUUCACCGCA---CACCGGUGCACUUAAAUUCACCACAGCCUUAAUAAUUCUUACGGC-AUUUCCGUGCUCAAUGGUGAAUGGUCGCACUGAA (((..((((((((((((............)))---)))))))))......((((((((.(((.............((((.-....)))))))...)))))))))))......... ( -40.81) >DroSec_CAF1 131095 111 + 1 ACCACGCACCGGUGUGCAAAAGAUUAACCGCA---CAUCGGUGCAUUUAAAUUCACCACAGCCGUAAUAAUUCCUACGGC-AUUUCUGUGCUCAAUGGUGAAAGGUCGCACUGAA (((..((((((((((((............)))---))))))))).......(((((((..((((((........))))))-.....((....)).))))))).)))......... ( -42.40) >DroSim_CAF1 122951 111 + 1 ACCACGCACCGGUGUGCAAAAGAUUCACCGCA---CAUCGGUGCAUUUAAAUUCACCACAGCCCUAAUAAUUCCUACGGC-AUUUCUGUGCUCAAUGGUGAAAGGUCGCACUGAA (((..((((((((((((............)))---))))))))).......(((((((..(((.((........)).)))-.....((....)).))))))).)))......... ( -36.70) >DroEre_CAF1 131436 111 + 1 ACCAUACACCGGUGUGCAAAAAAUUCACCGCA---CGCCGGUGCAUACAAAUUCACCACCGCCGUAAUGAUUCUUACGCG-AUUUUUGUACUUAAUGGCGACUGGUCACACUGAG ......(((((((((((............)))---))))))))...........((((.((((((((......))))(((-(...)))).......))))..))))......... ( -32.10) >DroYak_CAF1 130434 109 + 1 ACCACACACCGGUGUGCAAA-AAUUCACCGCA---CACCGGUAUAUUGAAAUUCACCACAGUCCUAAUGAUUC-UAUGAG-GUUUUUGUGCUUAAUGGCGACUGGCCACACUGAG .......((((((((((...-........)))---)))))))..........(((.(((((.(((.(((....-))).))-)...))))).....((((.....))))...))). ( -30.60) >DroPer_CAF1 150345 91 + 1 -----ACACCGGUGAGAGA---ACUCCCCACAGCAUGGUGGCCCAA-----UGCGC-----AAUUU-UUGCCCAAACGCGU-GUUCUCCUUCCG-CGGCGUGCGGCAACUCU--- -----.....((.(((...---.))).))...((((((....)).)-----)))..-----.....-(((((((..((((.-(........)))-))...)).)))))....--- ( -24.60) >consensus ACCACACACCGGUGUGCAAAAGAUUCACCGCA___CACCGGUGCAUUUAAAUUCACCACAGCCCUAAUAAUUCCUACGGC_AUUUCUGUGCUCAAUGGCGAAUGGUCACACUGAA .((..((((((((((((............)))...)))))))))........((((((................(((((......))))).....))))))..)).......... (-14.23 = -15.15 + 0.92)

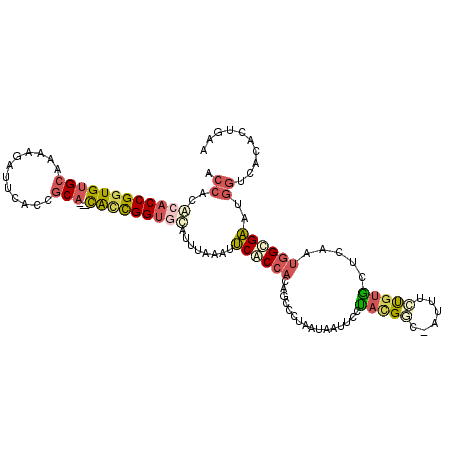
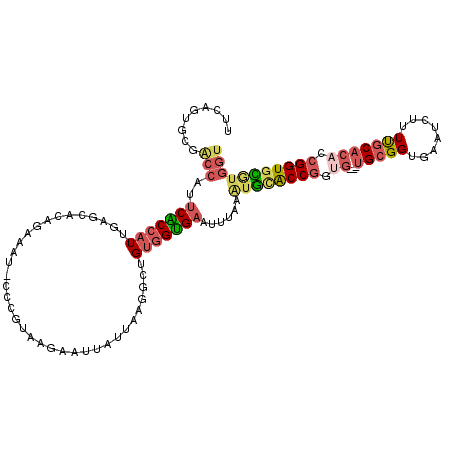
| Location | 16,676,356 – 16,676,467 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 69.30 |
| Mean single sequence MFE | -39.06 |
| Consensus MFE | -16.82 |
| Energy contribution | -17.87 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16676356 111 - 22407834 UUCAGUGCGACCAUUCACCAUUGAGCACGGAAAU-GCCGUAAGAAUUAUUAAGGCUGUGGUGAAUUUAAGUGCACCGGUG---UGCGGUGAAUCUUUUGCACACCGGUGCGUGGU .........((((((((((((..(((((((....-.))))((......))...))))))))))........(((((((((---(((((........))))))))))))))))))) ( -45.90) >DroSec_CAF1 131095 111 - 1 UUCAGUGCGACCUUUCACCAUUGAGCACAGAAAU-GCCGUAGGAAUUAUUACGGCUGUGGUGAAUUUAAAUGCACCGAUG---UGCGGUUAAUCUUUUGCACACCGGUGCGUGGU .........(((.((((((((.............-(((((((......))))))).)))))))).....((((((((.((---(((((........))))))).))))))))))) ( -43.04) >DroSim_CAF1 122951 111 - 1 UUCAGUGCGACCUUUCACCAUUGAGCACAGAAAU-GCCGUAGGAAUUAUUAGGGCUGUGGUGAAUUUAAAUGCACCGAUG---UGCGGUGAAUCUUUUGCACACCGGUGCGUGGU .........(((.((((((((.............-(((.(((......))).))).)))))))).....((((((((.((---(((((........))))))).))))))))))) ( -39.44) >DroEre_CAF1 131436 111 - 1 CUCAGUGUGACCAGUCGCCAUUAAGUACAAAAAU-CGCGUAAGAAUCAUUACGGCGGUGGUGAAUUUGUAUGCACCGGCG---UGCGGUGAAUUUUUUGCACACCGGUGUAUGGU ..(((..(.((((.(((((.....((........-.))((((......))))))))))))).)..)))((((((((((.(---(((((........)))))).)))))))))).. ( -40.10) >DroYak_CAF1 130434 109 - 1 CUCAGUGUGGCCAGUCGCCAUUAAGCACAAAAAC-CUCAUA-GAAUCAUUAGGACUGUGGUGAAUUUCAAUAUACCGGUG---UGCGGUGAAUU-UUUGCACACCGGUGUGUGGU ....(((((((.....))).....))))....((-(.....-(((((((((......))))))..))).(((((((((((---(((((......-.))))))))))))))))))) ( -37.30) >DroPer_CAF1 150345 91 - 1 ---AGAGUUGCCGCACGCCG-CGGAAGGAGAAC-ACGCGUUUGGGCAA-AAAUU-----GCGCA-----UUGGGCCACCAUGCUGUGGGGAGU---UCUCUCACCGGUGU----- ---...........((((((-.(...(((((((-.(.((..((.(((.-....)-----)).))-----.)).)((.((((...)))))).))---)))))..)))))))----- ( -28.60) >consensus UUCAGUGCGACCAUUCACCAUUGAGCACAGAAAU_CCCGUAAGAAUUAUUAAGGCUGUGGUGAAUUUAAAUGCACCGGUG___UGCGGUGAAUCUUUUGCACACCGGUGCGUGGU .........(((..(((((((...................................)))))))......((((((((.((...(((((........))))))).))))))))))) (-16.82 = -17.87 + 1.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:49 2006