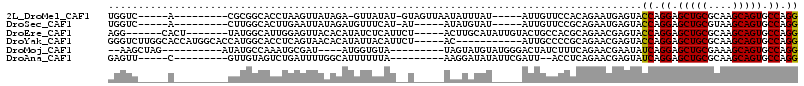
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
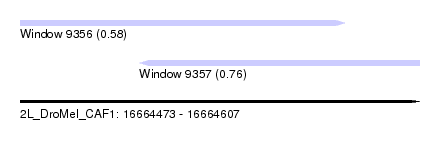
| Location | 16,664,473 – 16,664,607 |
| Length | 134 |
| Max. P | 0.760331 |

| Location | 16,664,473 – 16,664,582 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.37 |
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -26.22 |
| Energy contribution | -26.25 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
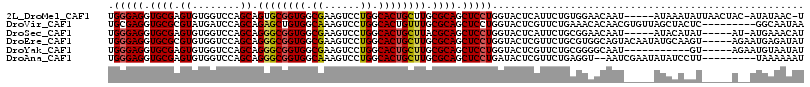
>2L_DroMel_CAF1 16664473 109 + 22407834 UGGGAGGUGCGAGUGUGGUCCAGCAGUGCGGUGGCGAAGUCCUGGCACUGCUUGCGCAGCUCCUGGUACUCAUUCUGUGGAACAAU-----AUAAAUAUUAACUAC-AUAUAAC-U .(((((.((((...(..((((((...(((....))).....)))).))..)...)))).)))))...........(((((...(((-----(....))))..))))-)......-. ( -32.20) >DroVir_CAF1 66853 107 + 1 UGCGAGGUGCGCGUAUGAUCCAGCAGAGCUGUGGCAAAGUCCUGGCACUGUUUGCGCAGCUCCUGGUACUCGUUCUGAAACACAACGUGUUAGCUACUC---------GGCAAUAA ((((((..((..(.((((.((((..((((((((.((((((....))....))))))))))))))))...)))).)...(((((...))))).))..)))---------.))).... ( -32.90) >DroSec_CAF1 119166 105 + 1 UGGGAGGUGCGAGUGUGGUCCAGCAGGGCGGUGGCGAAGUCCUGGCACUGCUUACGCAGCUCCUGGUACUCAUUCUGCGGAACAAU-----AUACAUAU-----AU-AUGAAACAU .(((((.((((..(((......)))((((((((.((......)).)))))))).)))).))))).(((....(((....)))....-----.)))....-----..-......... ( -30.00) >DroEre_CAF1 120110 111 + 1 UGGGAGGUGCGCGUGUGGUCCAGCAGGGCGGUGGCGAAGUCCUGGCACUGCUUGCGCAGCUCCUGGUACUCGUUCUGCGUGGCAGUACAAUAUGCAAGU-----AGAAUGAGAUAU .(((((.((((((.(..((....((((((.........))))))..))..).)))))).)))))....((((((((((...(((........)))..))-----)))))))).... ( -51.80) >DroYak_CAF1 119267 100 + 1 UGGGAGGUGCGAGUGUGGUCCAGCAGGGCGGUGGCGAAGUCCUGGCACUGCUUGCGCAGCUCCUGGUACUCGUUCUGCGGGGCAAU-----------GU-----AGAAUGUAAUAU .(((((.((((..(((......)))((((((((.((......)).)))))))).)))).)))))......(((((((((......)-----------))-----))))))...... ( -38.60) >DroAna_CAF1 61236 105 + 1 UGGGAGGUGCGAGUGUGGUCCAGCAGGGCGGUGGCAAAGUCCUGGCACUGCUUGCGCAGCUCCUGAUACUCGUUCUGAGGU--AAUCGAAUAUAUCCUU---------UAAAAAAU .(((((.((((..(((......)))((((((((.((......)).)))))))).)))).)))))(((.(((.....)))..--.)))............---------........ ( -32.90) >consensus UGGGAGGUGCGAGUGUGGUCCAGCAGGGCGGUGGCGAAGUCCUGGCACUGCUUGCGCAGCUCCUGGUACUCGUUCUGCGGAACAAU_____AUACACAU_____A__AUGAAAUAU .(((((.((((.((........)).((((((((.((......)).)))))))).)))).))))).................................................... (-26.22 = -26.25 + 0.03)
| Location | 16,664,513 – 16,664,607 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 59.11 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -13.82 |
| Energy contribution | -13.60 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
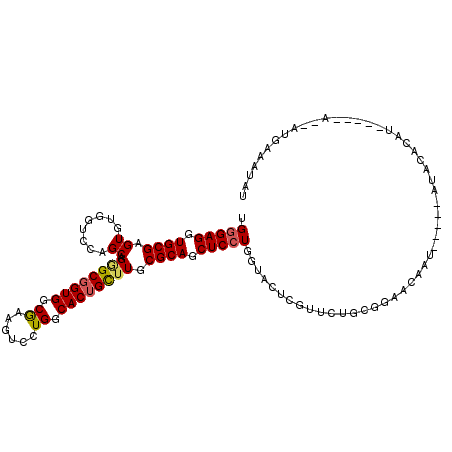
>2L_DroMel_CAF1 16664513 94 - 22407834 UGGUC-----A---------CGCGGCACCUAAGUUAUAGA-GUUAUAU-GUAGUUAAUAUUUAU-----AUUGUUCCACAGAAUGAGUACCAGGAGCUGCGCAAGCAGUGCCAGG .....-----.---------...(((.(((...(((((..-......)-))))....(((((((-----.(((.....))).)))))))..))).(((((....))))))))... ( -23.30) >DroSec_CAF1 119206 90 - 1 UGGUC-----A---------CUUGGCACUUGAAUUAUAGAUGUUUCAU-AU-----AUAUGUAU-----AUUGUUCCGCAGAAUGAGUACCAGGAGCUGCGUAAGCAGUGCCAGG .....-----.---------((((((.((((...((((.((((.....-..-----)))).)))-----)(..(((....)))..)....)))).(((((....))))))))))) ( -26.10) >DroEre_CAF1 120150 97 - 1 AGG------CACU-------UAUGGCAUUGGAGUUACACAUAUCUCAUUCU-----ACUUGCAUAUUGUACUGCCACGCAGAACGAGUACCAGGAGCUGCGCAAGCAGUGCCAGG .((------((((-------...(((....(((..........))).....-----.((((....((((.((((...)))).))))....)))).)))((....))))))))... ( -29.60) >DroYak_CAF1 119307 99 - 1 GGGUCUUGGCACCAUGGCACCAUGGCACCUCAGUAACACAUAUUACAUUCU-----AC-----------AUUGCCCCGCAGAACGAGUACCAGGAGCUGCGCAAGCAGUGCCAGG ....((((((((...(((.((.(((...(((.((((......)))).....-----..-----------.((((...))))...)))..))))).)))((....)).)))))))) ( -31.10) >DroMoj_CAF1 58642 90 - 1 --AAGCUAG----------AUAUGCCAAAUGCGAU----AUGGUGUA---------UAGUAUGUAUGGGACUAUCUUUCAGAACGAAUAUCAGGAGCUGCGAAAGCAGUGCCAGG --..(..((----------((((.(((.((((.((----((...)))---------).))))...))).).)))))..).............((.(((((....))))).))... ( -23.30) >DroAna_CAF1 61276 90 - 1 GAGUU-----C---------GUUGUAGUCUGAUUUUGGCAUUUUUUA---------AAGGAUAUAUUCGAUU--ACCUCAGAACGAGUAUCAGGAGCUGCGCAAGCAGUGCCAGG ..(((-----(---------(..((((((...((((((......)))---------)))((.....))))))--))..).))))........((.(((((....))))).))... ( -23.30) >consensus GGGUC_____A_________CAUGGCACCUGAGUUACACAUGUUUCAU__U_____AAAUAUAU_____AUUGUCCCGCAGAACGAGUACCAGGAGCUGCGCAAGCAGUGCCAGG .........................................................................................((.((.(((((....))))).)).)) (-13.82 = -13.60 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:45 2006