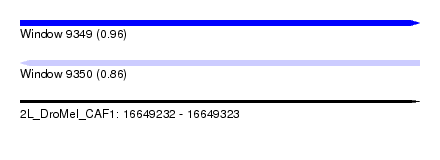
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,649,232 – 16,649,323 |
| Length | 91 |
| Max. P | 0.960442 |

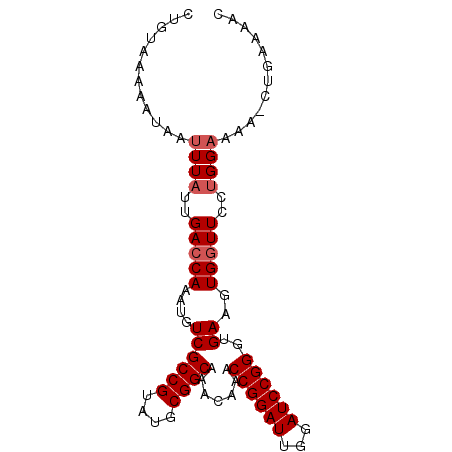
| Location | 16,649,232 – 16,649,323 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 94.74 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -20.44 |
| Energy contribution | -21.04 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16649232 91 + 22407834 CUGUAAAAAUAAUUUAUUGAACAAAUGUCGCCGUAUGCGGCAAACAAACCGGAUUGGAUCCGGGUGAAGUGGUUCCAGGAAAA-CUGAAAAC ..................((((.....((((((....)))).......((((((...))))))..))....))))(((.....-)))..... ( -22.60) >DroSec_CAF1 104029 91 + 1 CUGUAAAAAUAAUUUAUUGACCAAAUGUCGCCGUAUGCGGCAAACAAACCGGAUUGGAUCCGGGUGAAGUGGUUCCUGGAAAA-CUGAAAAC ............((((..(((((....((((((....)))).......((((((...))))))..))..)))))..))))...-........ ( -23.20) >DroSim_CAF1 106261 91 + 1 CUGUAAAAAUAAUUUAUUGACCAAAUGUCGCCGUAUGCGGCAAACAAACCGGAUUGGAUCCGGGUGAAGUGGUUCCUGGAAAA-CUGAAAAC ............((((..(((((....((((((....)))).......((((((...))))))..))..)))))..))))...-........ ( -23.20) >DroEre_CAF1 105313 90 + 1 CUGUGGAAAUAAUUUAUUGACCAAAUGUCGCCGUAUGCGGCAAACAAACCGGAUUGGAUCCGGGUGAAGUGGUUCCUGGUAAAUCAGAAA-- (((........((((((((((((....((((((....)))).......((((((...))))))..))..)))))...))))))))))...-- ( -24.00) >DroYak_CAF1 103232 89 + 1 CUGUAAAAAUAAUUUAUUGACCAAAUGUCGCCGUAUGCGGCAAACAAACCGGAUUGGAUCCGGUUGAAGUGGUUCCUGGAAAA-CUGUAA-- ............((((..(((((....((((((....)))).....((((((((...))))))))))..)))))..))))...-......-- ( -25.00) >consensus CUGUAAAAAUAAUUUAUUGACCAAAUGUCGCCGUAUGCGGCAAACAAACCGGAUUGGAUCCGGGUGAAGUGGUUCCUGGAAAA_CUGAAAAC ............((((..(((((....((((((....)))).......((((((...))))))..))..)))))..))))............ (-20.44 = -21.04 + 0.60)

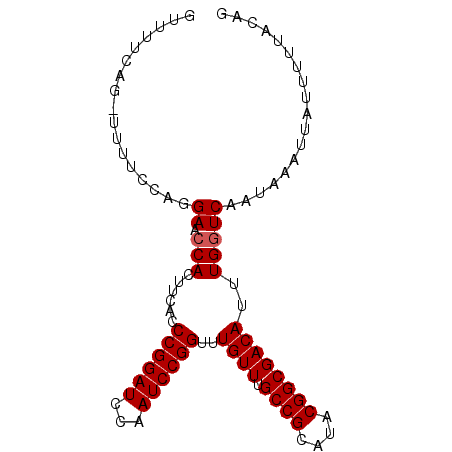
| Location | 16,649,232 – 16,649,323 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 94.74 |
| Mean single sequence MFE | -20.46 |
| Consensus MFE | -17.54 |
| Energy contribution | -17.74 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16649232 91 - 22407834 GUUUUCAG-UUUUCCUGGAACCACUUCACCCGGAUCCAAUCCGGUUUGUUUGCCGCAUACGGCGACAUUUGUUCAAUAAAUUAUUUUUACAG .....(((-.....)))((((........((((((...))))))..((((.((((....))))))))...)))).................. ( -19.70) >DroSec_CAF1 104029 91 - 1 GUUUUCAG-UUUUCCAGGAACCACUUCACCCGGAUCCAAUCCGGUUUGUUUGCCGCAUACGGCGACAUUUGGUCAAUAAAUUAUUUUUACAG .......(-((..(((((...........((((((...))))))..((((.((((....)))))))))))))..)))............... ( -19.50) >DroSim_CAF1 106261 91 - 1 GUUUUCAG-UUUUCCAGGAACCACUUCACCCGGAUCCAAUCCGGUUUGUUUGCCGCAUACGGCGACAUUUGGUCAAUAAAUUAUUUUUACAG .......(-((..(((((...........((((((...))))))..((((.((((....)))))))))))))..)))............... ( -19.50) >DroEre_CAF1 105313 90 - 1 --UUUCUGAUUUACCAGGAACCACUUCACCCGGAUCCAAUCCGGUUUGUUUGCCGCAUACGGCGACAUUUGGUCAAUAAAUUAUUUCCACAG --.(((((......)))))((((......((((((...))))))..((((.((((....))))))))..))))................... ( -22.00) >DroYak_CAF1 103232 89 - 1 --UUACAG-UUUUCCAGGAACCACUUCAACCGGAUCCAAUCCGGUUUGUUUGCCGCAUACGGCGACAUUUGGUCAAUAAAUUAUUUUUACAG --.....(-((..(((((.........((((((((...))))))))((((.((((....)))))))))))))..)))............... ( -21.60) >consensus GUUUUCAG_UUUUCCAGGAACCACUUCACCCGGAUCCAAUCCGGUUUGUUUGCCGCAUACGGCGACAUUUGGUCAAUAAAUUAUUUUUACAG .................((.(((......((((((...))))))..((((.((((....))))))))..))))).................. (-17.54 = -17.74 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:38 2006