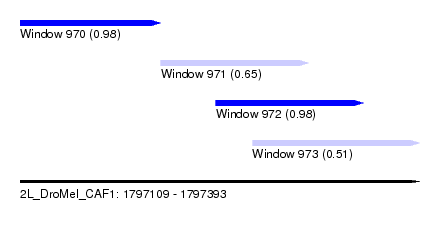
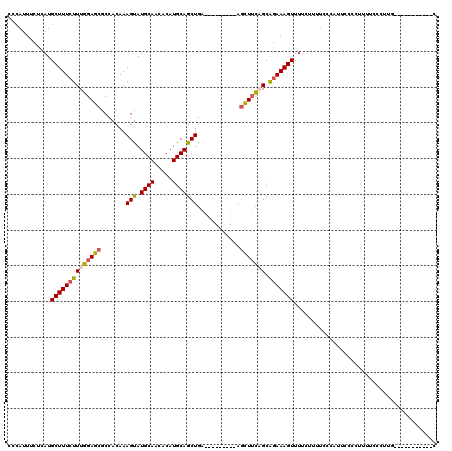
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,797,109 – 1,797,393 |
| Length | 284 |
| Max. P | 0.980466 |

| Location | 1,797,109 – 1,797,209 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.39 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -14.53 |
| Energy contribution | -14.66 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1797109 100 + 22407834 CCCAUUUCCCAUGCUUUCUUUGGAGCGCCACAAAGUAUGCAACACAUGCAGCUGA---------AGCUUCCGCAGAAAGUUUUCUUUUCCCAUUCCCCUUUUCCCUUA-----------C ............(((((((.(((((.((.....(((.((((.....)))))))..---------.))))))).)))))))............................-----------. ( -19.20) >DroSec_CAF1 4402 100 + 1 CCUAUUUCUCAUGCUUUCUUUGGAGCGCCACAAAGUAUGCAACACAUGCAGCUGA---------AGCUUCAGCAGAAAGUUUUCUUUUCCCAUUCCCCUUUUCCCUUU-----------C ............((((((((((((((.......(((.((((.....)))))))..---------.))))))).)))))))............................-----------. ( -19.90) >DroPer_CAF1 11137 114 + 1 GCCAUUUUUCAUGCUUUCUUUCGACCGCCGCAAAGCAUGCAACACAUGCAGCUGAGAGCUGGGAAAGUUGAGCGAAAAGUUUGC------CACUCCUCCAUCUCUGAGGGGGGAGAAACC ((.(((((((..((((.(((((..(((((....(((.((((.....)))))))..).)).)))))))..)))))))))))..))------..(((((((.((...)).)))))))..... ( -35.10) >DroEre_CAF1 6795 99 + 1 CCCAUUUUUCAUGCUUUCUUUGGAGCCCCACAAAGUAUGCAACACAUGCAGCUGA---------AGCU-CCGCAGAAAGUUUUCUUAUCCCGUUCCCCUUUUCCUUUG-----------C ............(((((((.((((((..((....(((((.....)))))...)).---------.)))-))).)))))))............................-----------. ( -21.50) >consensus CCCAUUUCUCAUGCUUUCUUUGGAGCGCCACAAAGUAUGCAACACAUGCAGCUGA_________AGCUUCAGCAGAAAGUUUUCUUUUCCCAUUCCCCUUUUCCCUUG___________C ............((((((((((((((.......(((.((((.....)))))))............))))))).)))))))........................................ (-14.53 = -14.66 + 0.13)

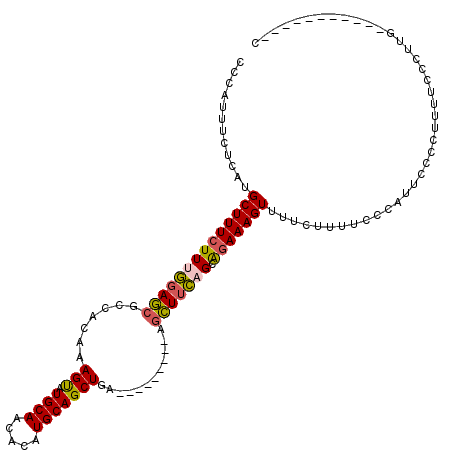
| Location | 1,797,209 – 1,797,314 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 94.95 |
| Mean single sequence MFE | -12.90 |
| Consensus MFE | -11.50 |
| Energy contribution | -11.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1797209 105 + 22407834 UCGCGUUAUAAUAAGUUACGCUUAGCAUUUUCUGUUUU-CCUUUCCGCAAAGUGCAUUUUUCACCUUUUAUCAUUAUUUCUUGGCUCAUUUUCCGAGCUUUUUCCC ..((((.((.....)).))))...(((((((.((....-......)).)))))))...........................(((((.......)))))....... ( -14.80) >DroSec_CAF1 4502 103 + 1 UCGCUUUAUAAUAAGUUACGCUUAGCAUUUUCUGUUUU-CCU-UCCGCAAAGUGCAUUUUUCACCUUUUAUCAUUAUUUCUU-GCUCAUUUUCCGAGCUUUUUUCC ...........((((.....))))(((((((.((....-...-..)).)))))))...........................-((((.......))))........ ( -11.70) >DroEre_CAF1 6894 104 + 1 UC-CUUCAUAAUAAGUUACGCUUAGCAUUUUCUGUUUUUCCUUUCCGCAAAGUGCAUUUUUCACCUUUUAUCAUUAUUUCUUGGCUCAUUUUCCGAGCUU-UUCCC ..-........((((.....))))(((((((.((...........)).)))))))...........................(((((.......))))).-..... ( -12.20) >consensus UCGCUUUAUAAUAAGUUACGCUUAGCAUUUUCUGUUUU_CCUUUCCGCAAAGUGCAUUUUUCACCUUUUAUCAUUAUUUCUUGGCUCAUUUUCCGAGCUUUUUCCC ...........((((.....))))(((((((.((...........)).)))))))............................((((.......))))........ (-11.50 = -11.50 + 0.00)

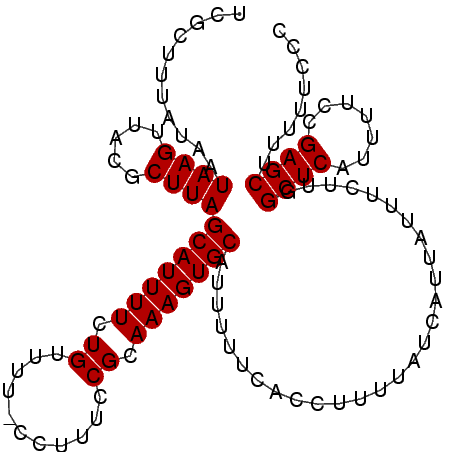
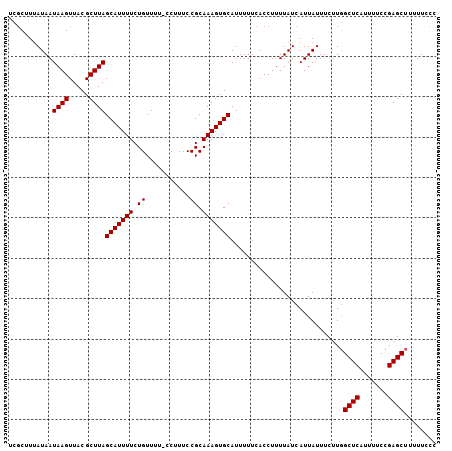
| Location | 1,797,248 – 1,797,353 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -15.24 |
| Energy contribution | -16.55 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1797248 105 + 22407834 CU--------------UUCCGCAAAGUGCAUUUUUCACCUUUUAUCAUUAUUUCUUGGCUCAUUUUCCGAGCUUUUUCCCUACCGAAGUUUCCCGACUUCGU-UCAAGGGGAUGAUGAUU ..--------------....((.....))............((((((((.......(((((.......)))))....((((..(((((((....))))))).-...)))))))))))).. ( -25.80) >DroSec_CAF1 4541 103 + 1 CU---------------UCCGCAAAGUGCAUUUUUCACCUUUUAUCAUUAUUUCUU-GCUCAUUUUCCGAGCUUUUUUCCUACCGAAGUUUCCCGACUUCGU-UCAAGGGGAUGAUGAUU ..---------------...((.....))..............((((((((..(((-((((.......))))...........(((((((....))))))).-...)))..)))))))). ( -25.00) >DroPer_CAF1 11289 119 + 1 CUGCCCCUGCCCCCCUCCCUUCUGAGUGCAUUUUUUACCUUUUAUCAUUAUUUGUUCGCACAUUUUCCUUGCUUU-UUCCAAAUGAAGCUUUCCAUCGAGAUGACGCGGGGAUGAUGAUU ....((((((....(((........((((............................)))).........(((((-........)))))........))).....))))))......... ( -20.59) >DroEre_CAF1 6933 103 + 1 CU--------------UUCCGCAAAGUGCAUUUUUCACCUUUUAUCAUUAUUUCUUGGCUCAUUUUCCGAGCUU-UUCCCUACCGA-GUUUCCCGACUCCGU-UCGAGGGGAUGAUGAUU ..--------------....((.....))............((((((((.......(((((.......))))).-..((((((.((-(((....))))).))-...)))))))))))).. ( -23.80) >consensus CU______________UUCCGCAAAGUGCAUUUUUCACCUUUUAUCAUUAUUUCUUGGCUCAUUUUCCGAGCUUUUUCCCUACCGAAGUUUCCCGACUUCGU_UCAAGGGGAUGAUGAUU ...........................................(((((((((((((.((((.......))))...........(((((((....))))))).....))))))))))))). (-15.24 = -16.55 + 1.31)

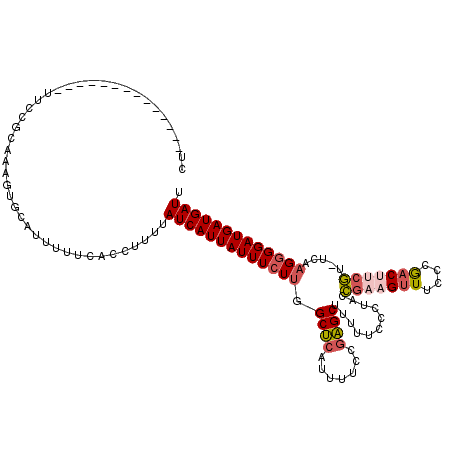
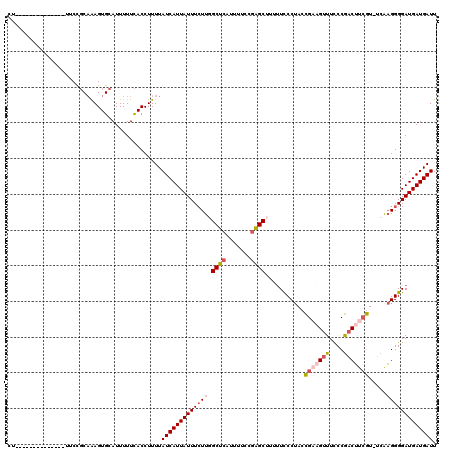
| Location | 1,797,274 – 1,797,393 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.87 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -19.64 |
| Energy contribution | -20.95 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1797274 119 + 22407834 UUUAUCAUUAUUUCUUGGCUCAUUUUCCGAGCUUUUUCCCUACCGAAGUUUCCCGACUUCGU-UCAAGGGGAUGAUGAUUGCUUUGGUCUAUGCAAAGUGUCUCAAUUUGGGUUCAUGUA .((((((((.......(((((.......)))))....((((..(((((((....))))))).-...))))))))))))((((..........)))).(((.(((.....)))..)))... ( -29.30) >DroSec_CAF1 4566 118 + 1 UUUAUCAUUAUUUCUU-GCUCAUUUUCCGAGCUUUUUUCCUACCGAAGUUUCCCGACUUCGU-UCAAGGGGAUGAUGAUUGCUUUGGUCUAUGCAAAGUGUCUCAAUUUGGGUUCACGUA ....(((((((..(((-((((.......))))...........(((((((....))))))).-...)))..)))))))((((..........)))).(((.(((.....)))..)))... ( -30.20) >DroPer_CAF1 11329 119 + 1 UUUAUCAUUAUUUGUUCGCACAUUUUCCUUGCUUU-UUCCAAAUGAAGCUUUCCAUCGAGAUGACGCGGGGAUGAUGAUUGCUUUGGUCUAUGCAAAGUGACUCAAUUUGGGUUCAUGUA ...((((((((((..((((.((((((....(((((-........)))))........))))))..)))))))))))))).((((((.......))))))(((((.....)))))...... ( -25.10) >DroEre_CAF1 6959 117 + 1 UUUAUCAUUAUUUCUUGGCUCAUUUUCCGAGCUU-UUCCCUACCGA-GUUUCCCGACUCCGU-UCGAGGGGAUGAUGAUUGCUUUGGUCUAUGCAAAGUGUCUCAAUUUGGCUUCAUGUA .((((((((.......(((((.......))))).-..((((((.((-(((....))))).))-...))))))))))))..((((((.......))))))(((.......)))........ ( -27.90) >consensus UUUAUCAUUAUUUCUUGGCUCAUUUUCCGAGCUUUUUCCCUACCGAAGUUUCCCGACUUCGU_UCAAGGGGAUGAUGAUUGCUUUGGUCUAUGCAAAGUGUCUCAAUUUGGGUUCAUGUA ...(((((((((((((.((((.......))))...........(((((((....))))))).....))))))))))))).((((((.......))))))..................... (-19.64 = -20.95 + 1.31)

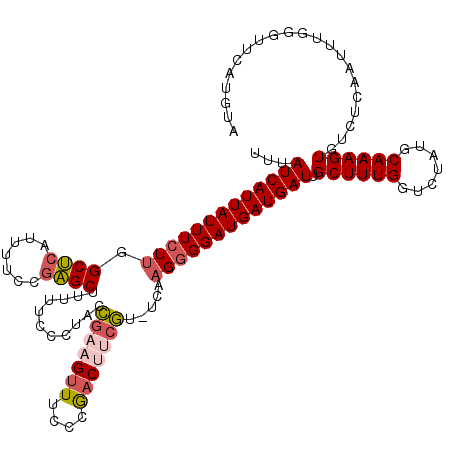
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:28 2006