
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,635,479 – 16,635,697 |
| Length | 218 |
| Max. P | 0.993647 |

| Location | 16,635,479 – 16,635,575 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 90.47 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -22.88 |
| Energy contribution | -23.16 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16635479 96 + 22407834 GCGAAUGUAUCUGUGACGCGUAUCUACAAUAUAUCUACAUGU--GUAUGUACACACAUAUCUUUUGUUGAGCGCGUAUCUCACGGAUGCAGUUCAAAU ..(((((((((((((((((((.((.((((.........((((--((......)))))).....)))).)))))))....))))))))))).))).... ( -30.84) >DroSec_CAF1 90253 93 + 1 GCGAAUGUAUCUGUGUCGCGUAUCU-----AUAUCUACAUGUGUGUAUGUACACACAUAUCUUUUGUCGAGCGCGUAUCUCACGGAUGCAGUUCAAAU ..(((((((((((((.(((((.((.-----(((.....((((((((....))))))))......))).))))))).....)))))))))).))).... ( -33.20) >DroSim_CAF1 92455 93 + 1 GCGAAUGUAUCUGUGUCGCGUAUCU-----AUAUCUACAUGUGUGUAUGUACACACAUAUCUUUUGUCGAGCGCGUAUCUCACGGAUGCAGUUCAAAU ..(((((((((((((.(((((.((.-----(((.....((((((((....))))))))......))).))))))).....)))))))))).))).... ( -33.20) >DroEre_CAF1 91447 91 + 1 GUGUAUGUAUCUGUGUCGCGUAUCU-----GCAUCUACAUGU--GUAUGUGCAUACAAAUCUUUUGUCGAGCGCGUAUCUCGCGGAUGCAGUUCAAAU .((..((((((((((.(((((.(((-----((((.(((....--))).))))).((((.....)))).))))))).....))))))))))...))... ( -29.00) >DroYak_CAF1 89331 91 + 1 GCGAAUGUAUCUGUGUCGCGUAUCU-----AUAUCUACACGU--GUCUGUACAUACAUCUCUUUUGUCGAGCGCGUAUCUCAGGGAUGCAGUUCAAAU ..((((((((((.((....(((...-----.....)))((((--(..((.(((...........)))))..)))))....)).))))))).))).... ( -21.60) >consensus GCGAAUGUAUCUGUGUCGCGUAUCU_____AUAUCUACAUGU__GUAUGUACACACAUAUCUUUUGUCGAGCGCGUAUCUCACGGAUGCAGUUCAAAU ..(((((((((((((.(((((.((...........((((((....))))))...(((.......))).))))))).....)))))))))).))).... (-22.88 = -23.16 + 0.28)



| Location | 16,635,575 – 16,635,666 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.87 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -18.95 |
| Energy contribution | -20.87 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
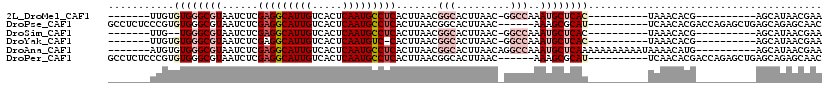
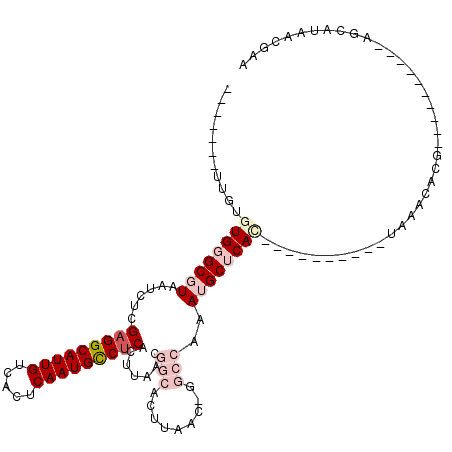
>2L_DroMel_CAF1 16635575 91 - 22407834 -------UUGUGUGGGCGUAAUCUCGAGGCAUUGUCACUCAAUGCCUCACUUAACGGCACUUAAC-GGCCAAAUGCUCAC----------UAAACACG----------AGCAUAACGAA -------....((((((((......(((((((((.....))))))))).......(((.......-.)))..))))))))----------........----------........... ( -31.00) >DroPse_CAF1 102623 103 - 1 GCCUCUCCCGUGUGGGCGUAAUCUCGAGGCAUUGUCACUCAAUGCCUCACUUAACGGCACUUAAC------AAAGCGCAU----------UCAACACGACCAGAGCUGAGCAGAGCAAC (((((((..((.(((.(((......(((((((((.....)))))))))........((.(((...------.))).))..----------.....))).)))..)).))).)).))... ( -32.00) >DroSim_CAF1 92548 89 - 1 -------UUG--UGGGCGUAAUCUCGAGGCAUUGUCACUCAAUGCCUCACUUAACGGCACUUAAC-GGCCAAAUGCUCAC----------UAAACACG----------AGCAUAACGAA -------..(--(((((((......(((((((((.....))))))))).......(((.......-.)))..))))))))----------........----------........... ( -31.00) >DroYak_CAF1 89422 90 - 1 -------UUGUGUGGGCGUAAUCUCGAGGCAUUGUCACUCAAUGUC-CACUUAACGGCACUUAAC-GGCCAAAUGCUCAC----------UAAACACG----------AGCAUAACGAA -------....((((((((......(.(((((((.....)))))))-).......(((.......-.)))..))))))))----------........----------........... ( -23.30) >DroAna_CAF1 31684 102 - 1 -------AUGUGUGGGCGUAAUCUCGAGGCAUUGUCACUCAAUGCCUCACUUAACGGCACUUAACAGGCCAAAUGCUCAAAAAAAAAAAUAAAACAUG----------AGCAUAACGAA -------.........(((......(((((((((.....))))))))).......(((.((....)))))..(((((((.................))----------))))).))).. ( -28.43) >DroPer_CAF1 103195 103 - 1 GCCUCUCCCGUGUGGGCGUAAUCUCGAGGCAUUGUCACUCAAUGCCUCACUUAACGGCACUUAAC------AAAGCGCAU----------UCAACACGACCAGAGCUGAGCAGAGCAAC (((((((..((.(((.(((......(((((((((.....)))))))))........((.(((...------.))).))..----------.....))).)))..)).))).)).))... ( -32.00) >consensus _______UUGUGUGGGCGUAAUCUCGAGGCAUUGUCACUCAAUGCCUCACUUAACGGCACUUAAC_GGCCAAAUGCUCAC__________UAAACACG__________AGCAUAACGAA ...........((((((((......(((((((((.....))))))))).......(((.........)))..))))))))....................................... (-18.95 = -20.87 + 1.92)
| Location | 16,635,595 – 16,635,697 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.58 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -14.70 |
| Energy contribution | -14.90 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
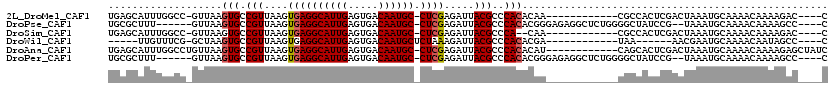
>2L_DroMel_CAF1 16635595 102 + 22407834 UGAGCAUUUGGCC-GUUAAGUGCCGUUAAGUGAGGCAUUGAGUGACAAUGC-CUCGAGAUUACGCCCACACAA------------CGCCACUCGACUAAAUGCAAAACAAAAGAC----C ...((((((((.(-(...((((.((((..(((((((((((.....))))))-)))..(........)))..))------------)).)))))).))))))))............----. ( -31.70) >DroPse_CAF1 102653 107 + 1 UGCGCUUU------GUUAAGUGCCGUUAAGUGAGGCAUUGAGUGACAAUGC-CUCGAGAUUACGCCCACACGGGAGAGGCUCUGGGGCUAUCCG--UAAAUGCAAAACAAAAGCC----C ...(((((------(((..(((.(((....((((((((((.....))))))-)))).....)))..)))(((((...((((....)))).))))--)........))).))))).----. ( -35.30) >DroSim_CAF1 92568 100 + 1 UGAGCAUUUGGCC-GUUAAGUGCCGUUAAGUGAGGCAUUGAGUGACAAUGC-CUCGAGAUUACGCCCA--CAA------------CGCCACUCGACUAAAUGCAAAACAAAAGAC----C ...((((((((.(-(...((((.((((...((((((((((.....))))))-)))).(........).--.))------------)).)))))).))))))))............----. ( -31.70) >DroWil_CAF1 183387 92 + 1 -----UUGUUUCG-GCUAAGUGCCGUUAAGUGAGGCAUUGAGUGACAAUGCUCUAAAGAUUACGCCCACACGA------------UAA------AACGAAUGCAAAACAAUAGCC----C -----((((((((-((.....))))....(((.(((...((((......))))....(....)))))))....------------...------..........)))))).....----. ( -19.30) >DroAna_CAF1 31714 107 + 1 UGAGCAUUUGGCCUGUUAAGUGCCGUUAAGUGAGGCAUUGAGUGACAAUGC-CUCGAGAUUACGCCCACACAU------------CAGCACUCGACUAAAUGCAAAACAAAAGAGCUAUC ...(((((((....((..((((((((....((((((((((.....))))))-)))).....))).........------------..)))))..)))))))))................. ( -31.10) >DroPer_CAF1 103225 107 + 1 UGCGCUUU------GUUAAGUGCCGUUAAGUGAGGCAUUGAGUGACAAUGC-CUCGAGAUUACGCCCACACGGGAGAGGCUCUGGGGCUAUCCG--UAAAUGCAAAACAAAAGCC----C ...(((((------(((..(((.(((....((((((((((.....))))))-)))).....)))..)))(((((...((((....)))).))))--)........))).))))).----. ( -35.30) >consensus UGAGCAUUUGGCC_GUUAAGUGCCGUUAAGUGAGGCAUUGAGUGACAAUGC_CUCGAGAUUACGCCCACACAA____________CGCCACUCGACUAAAUGCAAAACAAAAGAC____C ...................(((.(((....((((((((((.....)))))).)))).....)))..)))................................................... (-14.70 = -14.90 + 0.20)


| Location | 16,635,595 – 16,635,697 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.58 |
| Mean single sequence MFE | -34.35 |
| Consensus MFE | -13.95 |
| Energy contribution | -14.45 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16635595 102 - 22407834 G----GUCUUUUGUUUUGCAUUUAGUCGAGUGGCG------------UUGUGUGGGCGUAAUCUCGAG-GCAUUGUCACUCAAUGCCUCACUUAACGGCACUUAAC-GGCCAAAUGCUCA .----............((((((.((((((((.((------------(((.(((((......)))(((-((((((.....))))))))))).))))).))))...)-))).))))))... ( -38.90) >DroPse_CAF1 102653 107 - 1 G----GGCUUUUGUUUUGCAUUUA--CGGAUAGCCCCAGAGCCUCUCCCGUGUGGGCGUAAUCUCGAG-GCAUUGUCACUCAAUGCCUCACUUAACGGCACUUAAC------AAAGCGCA (----.(((((.(((.(((.....--.((((.(((((((((...)))...)).))))...)))).(((-((((((.....)))))))))........)))...)))------))))).). ( -36.50) >DroSim_CAF1 92568 100 - 1 G----GUCUUUUGUUUUGCAUUUAGUCGAGUGGCG------------UUG--UGGGCGUAAUCUCGAG-GCAUUGUCACUCAAUGCCUCACUUAACGGCACUUAAC-GGCCAAAUGCUCA .----............((((((.((((((((.((------------(((--.(((......)))(((-((((((.....)))))))))...))))).))))...)-))).))))))... ( -37.60) >DroWil_CAF1 183387 92 - 1 G----GGCUAUUGUUUUGCAUUCGUU------UUA------------UCGUGUGGGCGUAAUCUUUAGAGCAUUGUCACUCAAUGCCUCACUUAACGGCACUUAGC-CGAAACAA----- .----(((((........(((.((..------...------------.)).)))(.(((.......((.((((((.....))))))))......))).)...))))-).......----- ( -20.72) >DroAna_CAF1 31714 107 - 1 GAUAGCUCUUUUGUUUUGCAUUUAGUCGAGUGCUG------------AUGUGUGGGCGUAAUCUCGAG-GCAUUGUCACUCAAUGCCUCACUUAACGGCACUUAACAGGCCAAAUGCUCA .................((((((.(((((((((((------------((((....))))......(((-((((((.....)))))))))......))))))))....))).))))))... ( -35.90) >DroPer_CAF1 103225 107 - 1 G----GGCUUUUGUUUUGCAUUUA--CGGAUAGCCCCAGAGCCUCUCCCGUGUGGGCGUAAUCUCGAG-GCAUUGUCACUCAAUGCCUCACUUAACGGCACUUAAC------AAAGCGCA (----.(((((.(((.(((.....--.((((.(((((((((...)))...)).))))...)))).(((-((((((.....)))))))))........)))...)))------))))).). ( -36.50) >consensus G____GGCUUUUGUUUUGCAUUUAGUCGAGUGGCG____________UCGUGUGGGCGUAAUCUCGAG_GCAUUGUCACUCAAUGCCUCACUUAACGGCACUUAAC_GGCCAAAAGCUCA .................................................((((.(...(((....(((.((((((.....)))))))))..))).).))))................... (-13.95 = -14.45 + 0.50)
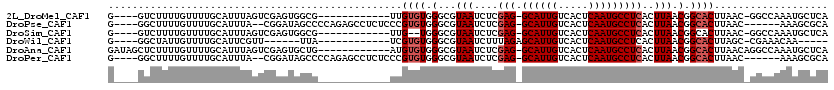

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:28 2006