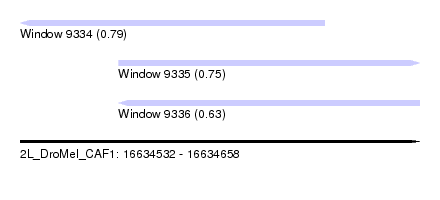
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,634,532 – 16,634,658 |
| Length | 126 |
| Max. P | 0.793652 |

| Location | 16,634,532 – 16,634,628 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 74.83 |
| Mean single sequence MFE | -19.46 |
| Consensus MFE | -11.88 |
| Energy contribution | -13.38 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16634532 96 - 22407834 CAUUU-CGCAGGC---GUGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCCUCCUUGCGCCCUC---------UUGCCCCUCACCGCACAUCGCUUUGCU .....-.((((((---(((((((.....)))))).......(((.((((..((..(((.......))).....---------..))...)))).)))...))).)))). ( -21.60) >DroSec_CAF1 89341 95 - 1 CAUUU-CGCAGGC---GUGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCCUCC-UGCGCCCUC---------UUGCCCCUCACCGCACUUCACUUUGCU .....-(((((((---(((((((.....))))))...)))....((((.....)))).....)-)))).....---------............(((........))). ( -21.40) >DroSim_CAF1 91529 95 - 1 CAUUU-CGCAGGC---GUGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCCUCC-UGCGCCCUC---------UUGCCCCUCACCGCACUUCACUUUGCU .....-(((((((---(((((((.....))))))...)))....((((.....)))).....)-)))).....---------............(((........))). ( -21.40) >DroEre_CAF1 90485 99 - 1 CAUUU-CGCAGGC---GUGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCCUCU-UGCGCCCCCUUCACCCCUUCACCCCUCACCGCACUUCAC-----U .....-.((.(((---(((((((.....))))))).((((.....))))..))).(((.....-.)))..........................)).......-----. ( -19.50) >DroWil_CAF1 182760 79 - 1 CAUUUUUGCGUGCAUUUUGUAUCUAAUAGAUACAUUUGGC-------------CACACCUAUC-UACCCCUUC---------GUGCCAUUCAAC--UCUCAGCC----- ...........((....((((((.....))))))..((((-------------.((.......-.........---------))))))......--.....)).----- ( -11.09) >DroYak_CAF1 88401 97 - 1 CAUUU-CGCAGGC---GGGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCCUCU-UGCGCCCC--UCGCCUCCUCGCCCCAUACCGCACUUCAC-----U .....-.((.(((---(((((((.....)))))...((((.....)))))))))..((.....-.(((....--.)))......))........)).......-----. ( -21.80) >consensus CAUUU_CGCAGGC___GUGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCCUCC_UGCGCCCUC_________UUGCCCCUCACCGCACUUCACU____U .......(((.((...(((((((.....)))))))...)).)))((((.....)))).................................................... (-11.88 = -13.38 + 1.50)

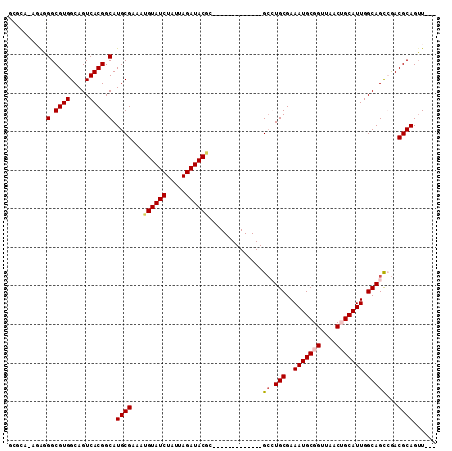
| Location | 16,634,563 – 16,634,658 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 84.39 |
| Mean single sequence MFE | -36.45 |
| Consensus MFE | -27.62 |
| Energy contribution | -27.98 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16634563 95 + 22407834 GCGCAAGGAGGGCGUGGCAGUCACGGCAUGCGAAAUGUAUCUAUUAGAUACAC-------------GCCUGCGAAAUGCGGUUAACUGCAUUGGCAGCCGACGCAGUU--- .(....)....(((((((.(((...(((.(((...((((((.....)))))))-------------)).)))..(((((((....)))))))))).))).))))....--- ( -37.00) >DroPse_CAF1 101790 102 + 1 GGGGA---GGGGCGUGGCAGUCACGGCAUGCGAAAUGUAUCUAUUAGAUACGA--GUGUGCG-AGUGCAUGCGAAAUGCGGUUAACAGCAUUGGCAGCAGACGCAGUU--- .....---...((((.((.(.(((.((((((....((((((.....)))))).--)))))).-.)))).(((..(((((........))))).)))))..))))....--- ( -33.10) >DroEre_CAF1 90520 94 + 1 GCGCA-AGAGGGCGUGGCAGUCACGGCAUGCGAAAUGUAUCUAUUAGAUACAC-------------GCCUGCGAAAUGCGGUUAACUGCAUUGGCAGCCGACGCAGUU--- .(...-.)...(((((((.(((...(((.(((...((((((.....)))))))-------------)).)))..(((((((....)))))))))).))).))))....--- ( -36.80) >DroYak_CAF1 88434 94 + 1 GCGCA-AGAGGGCGUGGCAGUCACGGCAUGCGAAAUGUAUCUAUUAGAUACCC-------------GCCUGCGAAAUGCGGUUAACUGCAUUGGCAGUCGACGCAGUU--- ((((.-...(((((((.(......).))))).....(((((.....)))))))-------------(.((((..(((((((....))))))).)))).)).)))....--- ( -33.70) >DroAna_CAF1 30888 110 + 1 GGAGA-AGGCGGCGUGGCAGUCACGGCAUGCGAAAUGUAUCUAUUAGAUACGCAAGUGUGCGAAGUGACUGCGAAAUGCGGUUAACUGCAUUGGCAGCCAACGCAGGUUGU .....-.(((.(((((((((((((.((((((....((((((.....))))))...))))))...))))))((..(((((((....))))))).)).))).))))..))).. ( -45.00) >DroPer_CAF1 102322 102 + 1 GGGGA---GGGGCGUGGCAGUCACGGCAUGCGAAAUGUAUCUAUUAGAUACGA--GUGUGCG-AGUGCAUGCGAAAUGCGGUUAACAGCAUUGGCAGCAGACGCAGUU--- .....---...((((.((.(.(((.((((((....((((((.....)))))).--)))))).-.)))).(((..(((((........))))).)))))..))))....--- ( -33.10) >consensus GCGCA_AGAGGGCGUGGCAGUCACGGCAUGCGAAAUGUAUCUAUUAGAUACGC_____________GCCUGCGAAAUGCGGUUAACUGCAUUGGCAGCCGACGCAGUU___ ..........(.((((.....)))).).((((...((((((.....))))))..............((.(((..(((((((....))))))).)))))...))))...... (-27.62 = -27.98 + 0.36)

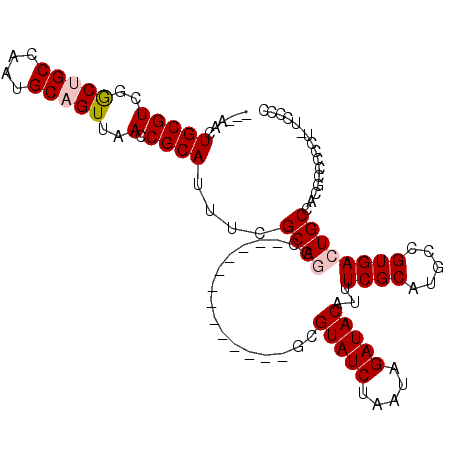
| Location | 16,634,563 – 16,634,658 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 84.39 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -20.91 |
| Energy contribution | -21.43 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16634563 95 - 22407834 ---AACUGCGUCGGCUGCCAAUGCAGUUAACCGCAUUUCGCAGGC-------------GUGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCCUCCUUGCGC ---...(((((..(((((....)))))..).))))...(((((((-------------(((((((.....)))))...((((.....)))).....))))).....)))). ( -30.60) >DroPse_CAF1 101790 102 - 1 ---AACUGCGUCUGCUGCCAAUGCUGUUAACCGCAUUUCGCAUGCACU-CGCACAC--UCGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCCC---UCCCC ---...((((..((((((.(((((.(....).)))))..))).)))..-))))...--..(((((.....)))))...........((((.....))))....---..... ( -25.60) >DroEre_CAF1 90520 94 - 1 ---AACUGCGUCGGCUGCCAAUGCAGUUAACCGCAUUUCGCAGGC-------------GUGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCCUCU-UGCGC ---...(((((..(((((....)))))..).))))...(((((((-------------(((((((.....)))))...((((.....)))).....)))))....-)))). ( -31.00) >DroYak_CAF1 88434 94 - 1 ---AACUGCGUCGACUGCCAAUGCAGUUAACCGCAUUUCGCAGGC-------------GGGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCCUCU-UGCGC ---...(((((..(((((....)))))..).))))...((((((.-------------(((((((.....)))))...........((((.....)))).)).))-)))). ( -31.50) >DroAna_CAF1 30888 110 - 1 ACAACCUGCGUUGGCUGCCAAUGCAGUUAACCGCAUUUCGCAGUCACUUCGCACACUUGCGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCGCCU-UCUCC ......((((((((((((....)))))))).))))....((((((((...(((....)))(((((.....)))))............))))))))..........-..... ( -35.90) >DroPer_CAF1 102322 102 - 1 ---AACUGCGUCUGCUGCCAAUGCUGUUAACCGCAUUUCGCAUGCACU-CGCACAC--UCGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCCC---UCCCC ---...((((..((((((.(((((.(....).)))))..))).)))..-))))...--..(((((.....)))))...........((((.....))))....---..... ( -25.60) >consensus ___AACUGCGUCGGCUGCCAAUGCAGUUAACCGCAUUUCGCAGGC_____________GCGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCCCCU_UCCCC ......(((((..(((((....)))))..).))))....((((.................(((((.....)))))...((((.....))))))))................ (-20.91 = -21.43 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:24 2006