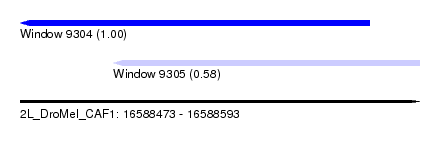
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,588,473 – 16,588,593 |
| Length | 120 |
| Max. P | 0.999955 |

| Location | 16,588,473 – 16,588,578 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 96.72 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -27.50 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.84 |
| SVM RNA-class probability | 0.999955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
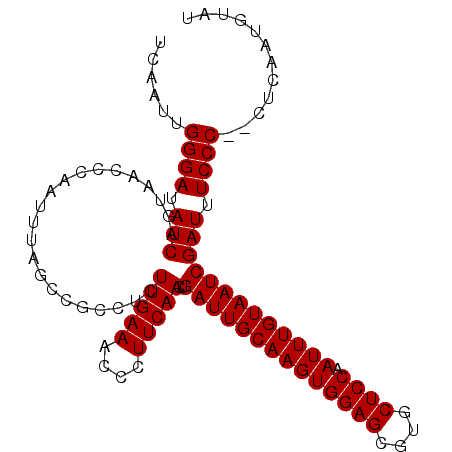
>2L_DroMel_CAF1 16588473 105 - 22407834 UCAAUUGGGAUAUCAGUAACCCAAUUUAGCCGCCUCUUGAAACCCUUCAACUGAUUGCAAGUGGAGCGUGCUCCAAUUUGUAAUCGAUUUCCCCCCUGAAUGUAU (((...((((.(((......................(((((....)))))..((((((((((((((....)))).))))))))))))).))))...)))...... ( -29.30) >DroSec_CAF1 43823 103 - 1 UCAAUUGGGAUAUCAGUAACCCAAUUUAGCCGCCUCUUGAAACCCUUCAACUGAUUGCAAGUGGAGCGUGCUCCAAUUUGUAAUCGAUUUCCC--CUCAAUGUAU ......((((.(((......................(((((....)))))..((((((((((((((....)))).))))))))))))).))))--.......... ( -28.00) >DroSim_CAF1 42632 103 - 1 UCAAUUGGGAUAUCAGUAACCCAAUUUAGCCGCCUCUUGAAACCCUUCAACUGAUUGCAAGUGGAGCGUGCUCCAAUUUGUAAUCGAUUUCCC--CUCAAUGUAU ......((((.(((......................(((((....)))))..((((((((((((((....)))).))))))))))))).))))--.......... ( -28.00) >DroEre_CAF1 42219 103 - 1 UAAAUUGGGAUAUCAGUAACCCAAUUUAGCCGCCCCUUGAAGCCCUUCAACUGAUUGCAAGUGGAGCGUGCUCCAAUUUGUAAUCGAUUUCCC--CUCAAUGCAU (((((((((.((....)).)))))))))...((...(((((....))))).(((((((((((((((....)))).))))))))))).......--......)).. ( -30.50) >DroYak_CAF1 42373 103 - 1 UCAAUUGGGAUAUCAGUAACCCAAUUUAGCGGCCCCUUGAAACCCUUCAACUGAUUGCAAGUGGAGCGUGCUCCAAUUUGUAAUCGAUUUCCC--CUCAAUGUAU ......((((.((((((...((........)).....((((....)))))))((((((((((((((....)))).))))))))))))).))))--.......... ( -28.10) >consensus UCAAUUGGGAUAUCAGUAACCCAAUUUAGCCGCCUCUUGAAACCCUUCAACUGAUUGCAAGUGGAGCGUGCUCCAAUUUGUAAUCGAUUUCCC__CUCAAUGUAU ......((((.(((......................(((((....)))))..((((((((((((((....)))).))))))))))))).))))............ (-27.50 = -27.50 + 0.00)

| Location | 16,588,501 – 16,588,593 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 95.40 |
| Mean single sequence MFE | -20.64 |
| Consensus MFE | -18.16 |
| Energy contribution | -18.56 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
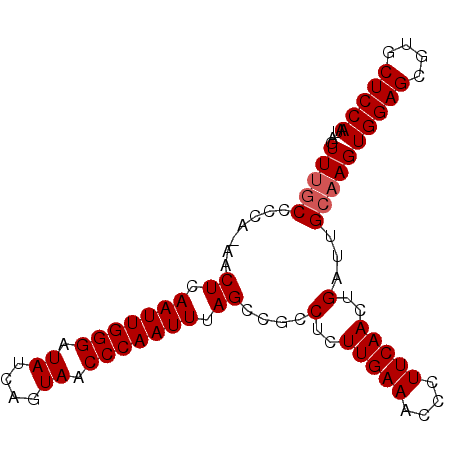
>2L_DroMel_CAF1 16588501 92 - 22407834 AGUUUGCCCCAAAACUCAAUUGGGAUAUCAGUAACCCAAUUUAGCCGCCUCUUGAAACCCUUCAACUGAUUGCAAGUGGAGCGUGCUCCAAU ..(((((.......((.(((((((.((....)).))))))).))....(..(((((....)))))..)...)))))(((((....))))).. ( -20.50) >DroSec_CAF1 43849 91 - 1 AGUUUGCCCCA-AACUCAAUUGGGAUAUCAGUAACCCAAUUUAGCCGCCUCUUGAAACCCUUCAACUGAUUGCAAGUGGAGCGUGCUCCAAU ..(((((....-..((.(((((((.((....)).))))))).))....(..(((((....)))))..)...)))))(((((....))))).. ( -20.50) >DroSim_CAF1 42658 91 - 1 AGUUUGCCCCA-AACUCAAUUGGGAUAUCAGUAACCCAAUUUAGCCGCCUCUUGAAACCCUUCAACUGAUUGCAAGUGGAGCGUGCUCCAAU ..(((((....-..((.(((((((.((....)).))))))).))....(..(((((....)))))..)...)))))(((((....))))).. ( -20.50) >DroEre_CAF1 42245 91 - 1 ACUUUCCCCCA-AACUAAAUUGGGAUAUCAGUAACCCAAUUUAGCCGCCCCUUGAAGCCCUUCAACUGAUUGCAAGUGGAGCGUGCUCCAAU ...........-...(((((((((.((....)).)))))))))(((((((((((.......((....))...)))).)).))).))...... ( -22.10) >DroYak_CAF1 42399 91 - 1 AGUUCGCCUUA-AACUCAAUUGGGAUAUCAGUAACCCAAUUUAGCGGCCCCUUGAAACCCUUCAACUGAUUGCAAGUGGAGCGUGCUCCAAU ...........-.....(((((((.((....)).)))))))..(((..(..(((((....)))))..)..)))...(((((....))))).. ( -19.60) >consensus AGUUUGCCCCA_AACUCAAUUGGGAUAUCAGUAACCCAAUUUAGCCGCCUCUUGAAACCCUUCAACUGAUUGCAAGUGGAGCGUGCUCCAAU ..(((((.......((.(((((((.((....)).))))))).))....(..(((((....)))))..)...)))))(((((....))))).. (-18.16 = -18.56 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:54 2006