
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,786,635 – 1,786,855 |
| Length | 220 |
| Max. P | 0.982243 |

| Location | 1,786,635 – 1,786,741 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 99.37 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -28.10 |
| Energy contribution | -28.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1786635 106 + 22407834 AUUAGUUCGCCGAAAUGCGCCAUCGCAUGCUCAUGAGGCAGACUCGUAAAUUCCAUCGAUAAUUUCCGCUAUCAUUCUGUUUUUGUCUGGCGGAAUAUUUAUUGGU ((.(((..(((...(((.((........)).)))..)))..))).)).......((((((((.((((((((.((.........))..))))))))...)))))))) ( -28.10) >DroSec_CAF1 43546 106 + 1 AUUAGUUCGCCGAAAUGCGCCAUCGCGUGCUCAUGAGGCAGACUCGUAAAUUCCAUCGAUAAUUUCCGCUAUCAUUCUGUUUUUGUCUGGCGGAAUAUUUAUUGGU ((.(((..(((.....((((......))))......)))..))).)).......((((((((.((((((((.((.........))..))))))))...)))))))) ( -28.60) >DroYak_CAF1 54775 106 + 1 AUUAGUUCGCCGAAAUGCGCCAUCGCAUGCUCAUGAGGCAGACUCGUAAAUUCCAUCGAUAAUUUCCGCUAUCAUUCUGUUUUUGUCUGGCGGAAUAUUUAUUGGU ((.(((..(((...(((.((........)).)))..)))..))).)).......((((((((.((((((((.((.........))..))))))))...)))))))) ( -28.10) >consensus AUUAGUUCGCCGAAAUGCGCCAUCGCAUGCUCAUGAGGCAGACUCGUAAAUUCCAUCGAUAAUUUCCGCUAUCAUUCUGUUUUUGUCUGGCGGAAUAUUUAUUGGU ((.(((..(((...(((.((........)).)))..)))..))).)).......((((((((.((((((((.((.........))..))))))))...)))))))) (-28.10 = -28.10 + -0.00)


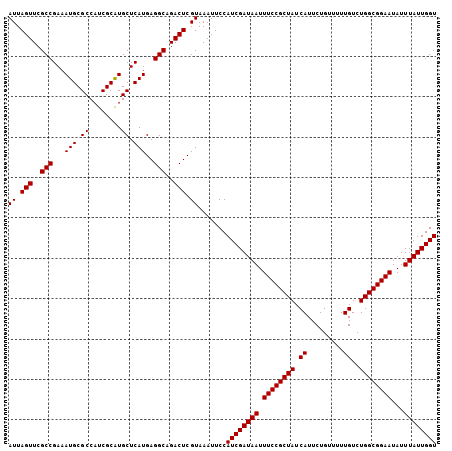
| Location | 1,786,675 – 1,786,781 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 98.11 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -28.90 |
| Energy contribution | -29.23 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1786675 106 + 22407834 GACUCGUAAAUUCCAUCGAUAAUUUCCGCUAUCAUUCUGUUUUUGUCUGGCGGAAUAUUUAUUGGUGCAGCCAUUAUUUUAUGGCAAUAUAAAAGUCACUCGGGGU ((((..((.....(((((((((.((((((((.((.........))..))))))))...)))))))))..(((((......)))))....))..))))......... ( -30.60) >DroSec_CAF1 43586 105 + 1 GACUCGUAAAUUCCAUCGAUAAUUUCCGCUAUCAUUCUGUUUUUGUCUGGCGGAAUAUUUAUUGGUACAGCCAU-AUUUUAUGGCAAUAUAAAAGUCACUCGGGGU ((((..((......((((((((.((((((((.((.........))..))))))))...))))))))...(((((-(...))))))....))..))))......... ( -28.00) >DroYak_CAF1 54815 106 + 1 GACUCGUAAAUUCCAUCGAUAAUUUCCGCUAUCAUUCUGUUUUUGUCUGGCGGAAUAUUUAUUGGUGCAGCCAUUAUUUUAUGGCAAUAUAAAAGUCAGUCGGGGU ((((..((.....(((((((((.((((((((.((.........))..))))))))...)))))))))..(((((......)))))....))..))))......... ( -30.60) >consensus GACUCGUAAAUUCCAUCGAUAAUUUCCGCUAUCAUUCUGUUUUUGUCUGGCGGAAUAUUUAUUGGUGCAGCCAUUAUUUUAUGGCAAUAUAAAAGUCACUCGGGGU ((((..((.....(((((((((.((((((((.((.........))..))))))))...)))))))))..(((((......)))))....))..))))......... (-28.90 = -29.23 + 0.33)


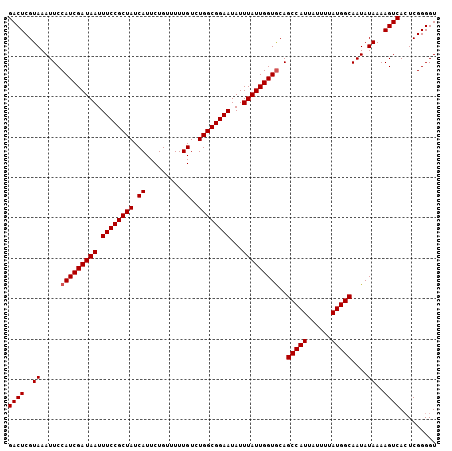
| Location | 1,786,741 – 1,786,855 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.40 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -25.68 |
| Energy contribution | -25.47 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1786741 114 + 22407834 GCAGCCAUUAUUUUAUGGCAAUAUAAAAGUCACUCGGGGUAGGGAGCGU------GUGUUUUAUUGGGCACCUACGUUAAAUUAUACGUAAUAGCUUUCCAAUUAUCCAAAUGACUUAGU ...(((((......))))).......((((((....((((((..((((.------((((((....)))))))(((((........)))))...)))..)....)))))...))))))... ( -32.50) >DroSec_CAF1 43652 113 + 1 ACAGCCAU-AUUUUAUGGCAAUAUAAAAGUCACUCGGGGUAGGGAGCGU------GUGUUUUAUUGGGCACCUACGUUAAAUUAUAUGUAAUAGCUUUCCAAUUAUCCAAAUGACUUAGU ...(((((-(...)))))).......((((((....((((((..((((.------((((((....)))))))(((((........)))))...)))..)....)))))...))))))... ( -30.40) >DroYak_CAF1 54881 120 + 1 GCAGCCAUUAUUUUAUGGCAAUAUAAAAGUCAGUCGGGGUAGGCAACAUAUACUCGUAUUUUAUUGGGCCCCUACAUUAAAUUACAUGUAAUAGCUUUCCAAUUAUCCAAAUGACUUAGU ...(((((......))))).......((((((....((((((....)...(((..(((.((((.((........)).)))).)))..))).............)))))...))))))... ( -27.20) >consensus GCAGCCAUUAUUUUAUGGCAAUAUAAAAGUCACUCGGGGUAGGGAGCGU______GUGUUUUAUUGGGCACCUACGUUAAAUUAUAUGUAAUAGCUUUCCAAUUAUCCAAAUGACUUAGU ...(((((......))))).......((((((....((((((((((((........)))))).((((((...(((((........)))))...))...))))))))))...))))))... (-25.68 = -25.47 + -0.22)



| Location | 1,786,741 – 1,786,855 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.40 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -24.65 |
| Energy contribution | -25.10 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1786741 114 - 22407834 ACUAAGUCAUUUGGAUAAUUGGAAAGCUAUUACGUAUAAUUUAACGUAGGUGCCCAAUAAAACAC------ACGCUCCCUACCCCGAGUGACUUUUAUAUUGCCAUAAAAUAAUGGCUGC ...(((((((((((......((..(((..((((((........))))))(((..........)))------..))).))....))))))))))).......(((((......)))))... ( -28.30) >DroSec_CAF1 43652 113 - 1 ACUAAGUCAUUUGGAUAAUUGGAAAGCUAUUACAUAUAAUUUAACGUAGGUGCCCAAUAAAACAC------ACGCUCCCUACCCCGAGUGACUUUUAUAUUGCCAUAAAAU-AUGGCUGU ...(((((((((((.....(((....)))................(((((.((............------..))..))))).))))))))))).......((((((...)-)))))... ( -25.34) >DroYak_CAF1 54881 120 - 1 ACUAAGUCAUUUGGAUAAUUGGAAAGCUAUUACAUGUAAUUUAAUGUAGGGGCCCAAUAAAAUACGAGUAUAUGUUGCCUACCCCGACUGACUUUUAUAUUGCCAUAAAAUAAUGGCUGC ...((((((.((((...(((((...(((.((((((........)))))).)))))))).........(((.....))).....)))).)))))).......(((((......)))))... ( -28.40) >consensus ACUAAGUCAUUUGGAUAAUUGGAAAGCUAUUACAUAUAAUUUAACGUAGGUGCCCAAUAAAACAC______ACGCUCCCUACCCCGAGUGACUUUUAUAUUGCCAUAAAAUAAUGGCUGC ...(((((((((((...(((((...((..((((((........))))))..)))))))...............(....)....))))))))))).......(((((......)))))... (-24.65 = -25.10 + 0.45)

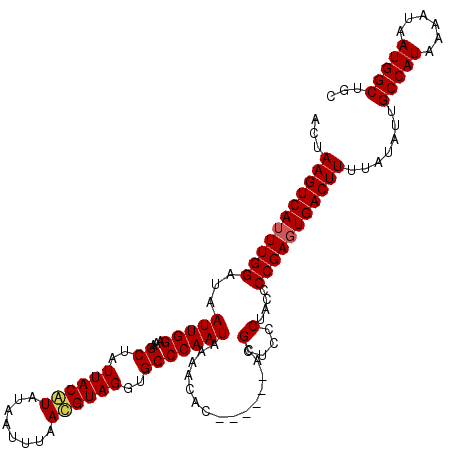

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:21 2006