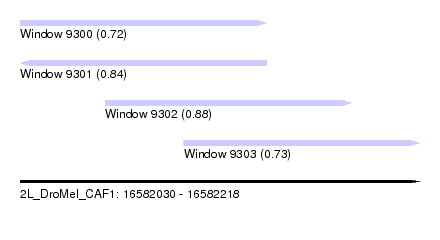
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,582,030 – 16,582,218 |
| Length | 188 |
| Max. P | 0.876807 |

| Location | 16,582,030 – 16,582,146 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -32.94 |
| Consensus MFE | -19.99 |
| Energy contribution | -20.43 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16582030 116 + 22407834 UUAUUAUCUCUUUCAAUCAGCCGCCUCCUCAUGUGGCAACCUUUGGGU--UUCGCAGACUUUCUGCACUCCCAACAAC-GUUGCU-CUGUCAUCGUUGUGGCAGUCGUGCUUAGGCAGCU ..................(((.((((...((((.((((((..(((((.--...((((.....))))...)))))....-))))))-(((((((....))))))).))))...)))).))) ( -37.30) >DroSec_CAF1 37287 116 + 1 UUAUUAUCUCUUUCAAUCAGCCGCCUCCUCAUGUGACAACCUUUGGGC--UUCGCAGACUUUCUGCACUCCCGACAAC-GUUGCU-CUGUCAUCGUUGUGGCAGUCGUGCUUAGGCAGCU ..................(((.((((...((((...((((..(((((.--...((((.....))))...)))))....-))))..-(((((((....))))))).))))...)))).))) ( -31.90) >DroSim_CAF1 35638 116 + 1 UUAUUAUCUCUUUCAAUCAGCCGCCUCCUCAUGUGACAACCUUUGGGC--UUCGCAGACUUUCUGCACUCCCGACAAC-GUUGCU-CUGUCAUCGUUGUGGCAGUCGUGCUUAGGCAGCU ..................(((.((((...((((...((((..(((((.--...((((.....))))...)))))....-))))..-(((((((....))))))).))))...)))).))) ( -31.90) >DroEre_CAF1 36083 118 + 1 CUAUUAUCUCUUUCAAUCAGCCGCCUCCUCAUGUGGCAAACUUAAGGUAUCUCGCAGACUUUCUGCAGAUCCGACAAC-GUUGCC-CUGUCACCGUUGUGGCAGUCGUGCUUAGGCAGCU ..................(((.((((...((((.(((((......((.((((.((((.....))))))))))......-.)))))-(((((((....))))))).))))...)))).))) ( -39.22) >DroWil_CAF1 41514 91 + 1 CCA--AUCUCUUUCAAUCUGCCGCUUCCCCA-----CAGCUCA---------CGAAG-----CUGCUCAUCUGACAACUGCUGC-----UUGUCGUUGUC---UUCGUGCUUAGGCAGCU ...--............((((((((((....-----.......---------.))))-----).((.(....((((((.((...-----..)).))))))---...).))...))))).. ( -23.70) >DroYak_CAF1 36340 117 + 1 CUAUUAUCUCUUUCAAUCAGCCGCCUCCUCAUGUGACUACCUUUGGGU--UUCGCAGACUUUCUGCAGUCCCGACAAC-GUUGCCCCUGUCAUCGUUGUGGCAGUCGUGCUUAGGCAGCU ..................(((.((((...(((..((((.((..((((.--...((((.....))))...))))(((((-(.(((....).)).)))))))).)))))))...)))).))) ( -33.60) >consensus CUAUUAUCUCUUUCAAUCAGCCGCCUCCUCAUGUGACAACCUUUGGGU__UUCGCAGACUUUCUGCACUCCCGACAAC_GUUGCU_CUGUCAUCGUUGUGGCAGUCGUGCUUAGGCAGCU ...................((.((((...((((....................((((.....))))....((.(((((.((.((....)).)).)))))))....))))...)))).)). (-19.99 = -20.43 + 0.45)


| Location | 16,582,030 – 16,582,146 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -39.28 |
| Consensus MFE | -21.80 |
| Energy contribution | -22.08 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16582030 116 - 22407834 AGCUGCCUAAGCACGACUGCCACAACGAUGACAG-AGCAAC-GUUGUUGGGAGUGCAGAAAGUCUGCGAA--ACCCAAAGGUUGCCACAUGAGGAGGCGGCUGAUUGAAAGAGAUAAUAA ((((((((...((.(.(((.((......)).)))-.(((((-....(((((..(((((.....)))))..--.)))))..)))))..).))...)))))))).................. ( -37.90) >DroSec_CAF1 37287 116 - 1 AGCUGCCUAAGCACGACUGCCACAACGAUGACAG-AGCAAC-GUUGUCGGGAGUGCAGAAAGUCUGCGAA--GCCCAAAGGUUGUCACAUGAGGAGGCGGCUGAUUGAAAGAGAUAAUAA ((((((((...((.(((.(((((((((.((....-..)).)-))))).(((..(((((.....)))))..--.)))...))).)))...))...)))))))).................. ( -42.70) >DroSim_CAF1 35638 116 - 1 AGCUGCCUAAGCACGACUGCCACAACGAUGACAG-AGCAAC-GUUGUCGGGAGUGCAGAAAGUCUGCGAA--GCCCAAAGGUUGUCACAUGAGGAGGCGGCUGAUUGAAAGAGAUAAUAA ((((((((...((.(((.(((((((((.((....-..)).)-))))).(((..(((((.....)))))..--.)))...))).)))...))...)))))))).................. ( -42.70) >DroEre_CAF1 36083 118 - 1 AGCUGCCUAAGCACGACUGCCACAACGGUGACAG-GGCAAC-GUUGUCGGAUCUGCAGAAAGUCUGCGAGAUACCUUAAGUUUGCCACAUGAGGAGGCGGCUGAUUGAAAGAGAUAAUAG ((((((((...((.(.(((.(((....))).)))-(((((.-.(((..((((((((((.....)))).)))).)).)))..))))).).))...)))))))).................. ( -39.50) >DroWil_CAF1 41514 91 - 1 AGCUGCCUAAGCACGAA---GACAACGACAA-----GCAGCAGUUGUCAGAUGAGCAG-----CUUCG---------UGAGCUG-----UGGGGAAGCGGCAGAUUGAAAGAGAU--UGG ..(((((...((.....---((((((..(..-----...)..))))))......((((-----(((..---------.))))))-----)......)))))))............--... ( -25.50) >DroYak_CAF1 36340 117 - 1 AGCUGCCUAAGCACGACUGCCACAACGAUGACAGGGGCAAC-GUUGUCGGGACUGCAGAAAGUCUGCGAA--ACCCAAAGGUAGUCACAUGAGGAGGCGGCUGAUUGAAAGAGAUAAUAG ((((((((...((.(((((((........(((((.(....)-.)))))(((..(((((.....)))))..--.)))...)))))))...))...)))))))).................. ( -47.40) >consensus AGCUGCCUAAGCACGACUGCCACAACGAUGACAG_AGCAAC_GUUGUCGGGAGUGCAGAAAGUCUGCGAA__ACCCAAAGGUUGUCACAUGAGGAGGCGGCUGAUUGAAAGAGAUAAUAA ((((((((...((............(((((((..........))))))).....((((.....))))......................))...)))))))).................. (-21.80 = -22.08 + 0.28)

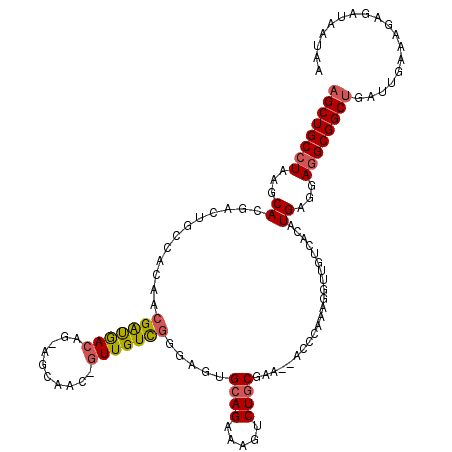
| Location | 16,582,070 – 16,582,186 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.62 |
| Mean single sequence MFE | -35.45 |
| Consensus MFE | -28.14 |
| Energy contribution | -29.10 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16582070 116 + 22407834 CUUUGGGU--UUCGCAGACUUUCUGCACUCCCAACAACGUUGCU-CUGUCAUCGUUGUGGCAGUCGUGCUUAGGCAGCUAUCAUAAAUUGUUGCUUAAAUUGCAUAUCAUAUGGUGACA ..(((((.--...((((.....))))...)))))....((..((-(((((((....)))))))..(((((((((((((...........)))))))))...)))).......))..)). ( -36.70) >DroSec_CAF1 37327 116 + 1 CUUUGGGC--UUCGCAGACUUUCUGCACUCCCGACAACGUUGCU-CUGUCAUCGUUGUGGCAGUCGUGCUUAGGCAGCUAUCAUAAAUUGUUGCUUAAAUUGCAUAUCAUACGGUGACA ..(((((.--...((((.....))))...)))))..........-..((((((((((..(((((.....(((((((((...........))))))))))))))....)).)))))))). ( -36.20) >DroSim_CAF1 35678 116 + 1 CUUUGGGC--UUCGCAGACUUUCUGCACUCCCGACAACGUUGCU-CUGUCAUCGUUGUGGCAGUCGUGCUUAGGCAGCUAUCAUAAAUUGUUGCUUAAAUUGCAUAUCAUACGGUGACA ..(((((.--...((((.....))))...)))))..........-..((((((((((..(((((.....(((((((((...........))))))))))))))....)).)))))))). ( -36.20) >DroEre_CAF1 36123 118 + 1 CUUAAGGUAUCUCGCAGACUUUCUGCAGAUCCGACAACGUUGCC-CUGUCACCGUUGUGGCAGUCGUGCUUAGGCAGCUAUCAUAAAUUGUUGCUUAAAUUGCAUAUCAUACGGUGACG .....((.((((.((((.....)))))))))).....(((..((-(((((((....)))))))..(((((((((((((...........)))))))))...)))).......))..))) ( -41.80) >DroYak_CAF1 36380 117 + 1 CUUUGGGU--UUCGCAGACUUUCUGCAGUCCCGACAACGUUGCCCCUGUCAUCGUUGUGGCAGUCGUGCUUAGGCAGCUAUCAUAAAUUGUUGCUUAAAUUGCAUAUCAUACUGUGACA ..(((((.--...((((.....))))...)))))....((..(..(((((((....)))))))..(((((((((((((...........)))))))))...))))........)..)). ( -35.50) >DroPer_CAF1 45440 87 + 1 CUCUGG--------------------AG--------AGCUUGCC-CCGUCAUCGUUGUU---UUCGUGCUUAGGCAGCUAUCAUAAAUUGUUGCUUAAAUUGCAUAUCACACGAUGACA (((...--------------------.)--------))......-..((((((((.((.---...(((((((((((((...........)))))))))...))))...)))))))))). ( -26.30) >consensus CUUUGGGU__UUCGCAGACUUUCUGCACUCCCGACAACGUUGCC_CUGUCAUCGUUGUGGCAGUCGUGCUUAGGCAGCUAUCAUAAAUUGUUGCUUAAAUUGCAUAUCAUACGGUGACA ..(((((......((((.....))))...))))).............((((((((.((((.....(((((((((((((...........)))))))))...)))).)))))))))))). (-28.14 = -29.10 + 0.96)
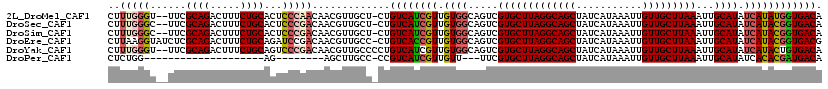

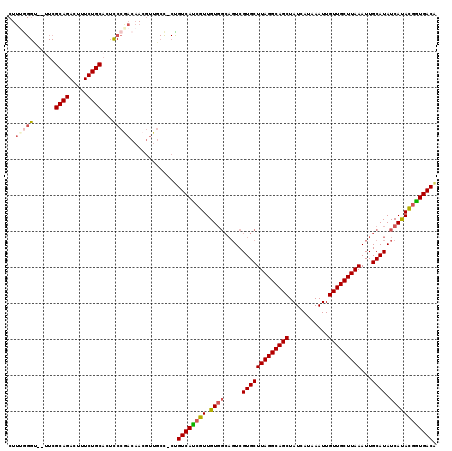
| Location | 16,582,107 – 16,582,218 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 85.81 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -24.65 |
| Energy contribution | -25.15 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16582107 111 + 22407834 UUGCU-CUGUCAUCGUUGUGGCAGUCGUGCUUAGGCAGCUAUCAUAAAUUGUUGCUUAAAUUGCAUAUCAUAUGGUGACAUCGAGUGGCGACGUU---CCUUCCUUUUUCCCCAC (..((-((((((((((((..(((((.....(((((((((...........))))))))))))))....)).)))))))))..)))..).......---................. ( -29.00) >DroPse_CAF1 46228 110 + 1 UUGCC-CCGUCAUCGUUGUU---UUCGUGCUUAGGCAGCUAUCAUAAAUUGUUGCUUAAAUUGCAUAUCACACGAUGACAUCAAAUGACAACGCUGAGCCCUCGAU-CUGCCCAC .....-..((((((((.((.---...(((((((((((((...........)))))))))...))))...)))))))))).............((.((........)-).)).... ( -27.30) >DroSim_CAF1 35715 110 + 1 UUGCU-CUGUCAUCGUUGUGGCAGUCGUGCUUAGGCAGCUAUCAUAAAUUGUUGCUUAAAUUGCAUAUCAUACGGUGACAUCGAGUGGCAACGUC---CCUUCCUU-UUCCCCAC (..((-((((((((((((..(((((.....(((((((((...........))))))))))))))....)).)))))))))..)))..).......---........-........ ( -30.80) >DroEre_CAF1 36162 110 + 1 UUGCC-CUGUCACCGUUGUGGCAGUCGUGCUUAGGCAGCUAUCAUAAAUUGUUGCUUAAAUUGCAUAUCAUACGGUGACGUCGAGUGGCAACGUU---CCUUCCUU-UUUUCCAU (((((-((((((((((((..(((((.....(((((((((...........))))))))))))))....)).))))))))....)).)))))....---........-........ ( -33.40) >DroYak_CAF1 36417 111 + 1 UUGCCCCUGUCAUCGUUGUGGCAGUCGUGCUUAGGCAGCUAUCAUAAAUUGUUGCUUAAAUUGCAUAUCAUACUGUGACAUCGAGUGGCAUCGUC---CCUUCCUU-UUACUCAU .......((((((((.(((.(((((.(((((((((((((...........)))))))))...)))).....))))).))).))).))))).....---........-........ ( -29.80) >DroPer_CAF1 45451 110 + 1 UUGCC-CCGUCAUCGUUGUU---UUCGUGCUUAGGCAGCUAUCAUAAAUUGUUGCUUAAAUUGCAUAUCACACGAUGACAUCAAAUGACAACGCUGAGCCCUCGAU-CUGCCCAC .....-..((((((((.((.---...(((((((((((((...........)))))))))...))))...)))))))))).............((.((........)-).)).... ( -27.30) >consensus UUGCC_CUGUCAUCGUUGUGGCAGUCGUGCUUAGGCAGCUAUCAUAAAUUGUUGCUUAAAUUGCAUAUCAUACGGUGACAUCGAGUGGCAACGUU___CCUUCCUU_UUCCCCAC (((((..(((((((((.((((.....(((((((((((((...........)))))))))...)))).)))))))))))))......)))))........................ (-24.65 = -25.15 + 0.50)


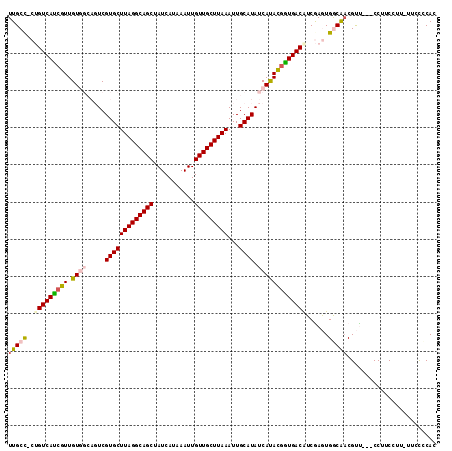
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:52 2006