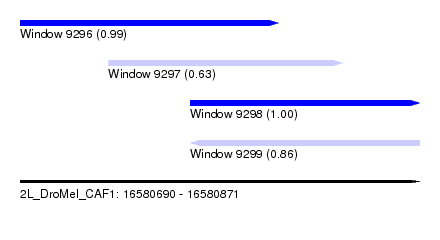
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,580,690 – 16,580,871 |
| Length | 181 |
| Max. P | 0.997784 |

| Location | 16,580,690 – 16,580,807 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 93.36 |
| Mean single sequence MFE | -31.26 |
| Consensus MFE | -28.86 |
| Energy contribution | -30.14 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16580690 117 + 22407834 AGUUGAUGGCAGUGGUUUGGUCGAAAAAGAGCUCAUUCCUGCUCAAGGUAUUUUCGACAAUGACAAUAA-AAAAAUUGCUGGCUCUAAAGUUGAGCCGGACUAAAAAUAAAGUUUGGC ....((.(.(((..((((.((((((((.((((........))))......))))))))...........-..))))..))).))).........((((((((........)))))))) ( -31.24) >DroSec_CAF1 35946 117 + 1 AGUUGGUGGCAGUGGUUCGGUCAGAAAGGAGCUCAUUCCUGCUCAAGGUAUUUUCGACAAUGACAAUAA-AAAAAUUGCUGGCUCUAAAGUUGAGCCGGACUAAAAAUAAAGUUUGGC ..((((.(.(((..(((..((((.....((((........))))..((.....)).....)))).....-...)))..))).).))))......((((((((........)))))))) ( -29.10) >DroSim_CAF1 33896 117 + 1 AGUUGGUGGCAGUGGUUCGGUCAGAAAGGAGCUCAUUCCUGCUCAAGGUAUUUUCGACAAUGACAAUAA-AAAAAUUGCUGGCUCUAAAGUUGAGCCGGACUAAAAAUAAAGUUUGGC ..((((.(.(((..(((..((((.....((((........))))..((.....)).....)))).....-...)))..))).).))))......((((((((........)))))))) ( -29.10) >DroEre_CAF1 34754 118 + 1 AGUUGGGGGCAGUGGUUUGGUAGAAAAGGAGCUCAUUCUUGCUCAAGGUAUUUUCGACAAUGAUAAUAAGAAAAAUUGCUGGCUCCAGAGUUAAGCCGGACUAAAAAUAAAGUUUGGC ..((((((.(((..((((.((.(((((.((((........))))......))))).))..............))))..))).))))))......((((((((........)))))))) ( -35.66) >DroYak_CAF1 35026 116 + 1 AGUUGG-GGCAGUGGCUUGGUAGAAAAGGAGCUCAUUCUUGCUCAAGGUAUUUUCGACAAUGUCAAUAA-AAAAAUUGCUGGCUCUACAGUUAAGCCGGACUAAAAAUAAAGUUUGGC .((.((-(.((((((((((...(((((.((((........))))......)))))..))).)))(((..-....))))))).))).))......((((((((........)))))))) ( -31.20) >consensus AGUUGGUGGCAGUGGUUUGGUCGAAAAGGAGCUCAUUCCUGCUCAAGGUAUUUUCGACAAUGACAAUAA_AAAAAUUGCUGGCUCUAAAGUUGAGCCGGACUAAAAAUAAAGUUUGGC ..((((.(.(((..((((.((((((((.((((........))))......))))))))..............))))..))).).))))......((((((((........)))))))) (-28.86 = -30.14 + 1.28)

| Location | 16,580,730 – 16,580,836 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 95.39 |
| Mean single sequence MFE | -24.11 |
| Consensus MFE | -19.86 |
| Energy contribution | -20.26 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16580730 106 + 22407834 GCUCAAGGUAUUUUCGACAAUGACAAUAA-AAAAAUUGCUGGCUCUAAAGUUGAGCCGGACUAAAAAUAAAGUUUGGCCUUGUCAGCUUUAAGUUAAUCCACGAAAG ...........(((((.......((((..-....)))).(((((.(((((((((((((((((........))))))))....)))))))))))))).....))))). ( -25.60) >DroSec_CAF1 35986 106 + 1 GCUCAAGGUAUUUUCGACAAUGACAAUAA-AAAAAUUGCUGGCUCUAAAGUUGAGCCGGACUAAAAAUAAAGUUUGGCCUUGUCAGCUUUAAGUUAAUCCACGAAAG ...........(((((.......((((..-....)))).(((((.(((((((((((((((((........))))))))....)))))))))))))).....))))). ( -25.60) >DroSim_CAF1 33936 106 + 1 GCUCAAGGUAUUUUCGACAAUGACAAUAA-AAAAAUUGCUGGCUCUAAAGUUGAGCCGGACUAAAAAUAAAGUUUGGCCUUGUCAGCUUUAAGUUAAUCCACGAAAG ...........(((((.......((((..-....)))).(((((.(((((((((((((((((........))))))))....)))))))))))))).....))))). ( -25.60) >DroEre_CAF1 34794 107 + 1 GCUCAAGGUAUUUUCGACAAUGAUAAUAAGAAAAAUUGCUGGCUCCAGAGUUAAGCCGGACUAAAAAUAAAGUUUGGCCUUGUCAGCUUUAAGUUAAUCCAGAGAAG .(((..(((.(((((..............)))))...((((((....(((....((((((((........))))))))))))))))).........)))..)))... ( -22.04) >DroYak_CAF1 35065 106 + 1 GCUCAAGGUAUUUUCGACAAUGUCAAUAA-AAAAAUUGCUGGCUCUACAGUUAAGCCGGACUAAAAAUAAAGUUUGGCCUUGUCAGCUUUAAGUUUAUCCAGGGAAG .(((..(((......(((...))).....-.......((((......))))...)))(((.((((..(((((((.(((...))))))))))..))))))).)))... ( -21.70) >consensus GCUCAAGGUAUUUUCGACAAUGACAAUAA_AAAAAUUGCUGGCUCUAAAGUUGAGCCGGACUAAAAAUAAAGUUUGGCCUUGUCAGCUUUAAGUUAAUCCACGAAAG (((((((((........((((.............))))(((((((.......))))))).................))))))..))).................... (-19.86 = -20.26 + 0.40)

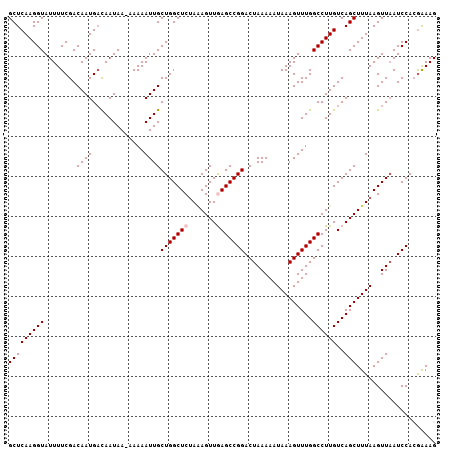
| Location | 16,580,767 – 16,580,871 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 82.04 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -20.85 |
| Energy contribution | -21.85 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
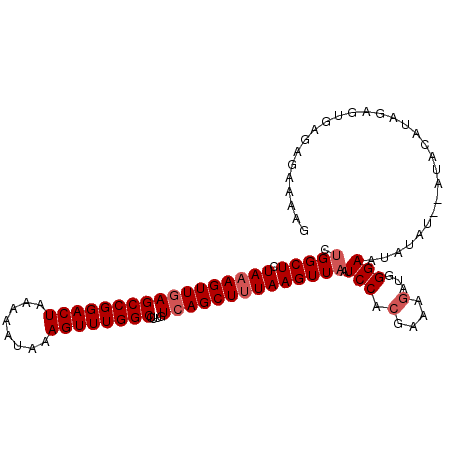
>2L_DroMel_CAF1 16580767 104 + 22407834 CUGGCUCUAAAGUUGAGCCGGACUAAAAAUAAAGUUUGGCCUUGUCAGCUUUAAGUUAAUCCACGAAAGAUGGGAAUAUAUGUAUAUAUAGAAGGAGAGAAAAG .(((((.(((((((((((((((((........))))))))....)))))))))))))).(((((....).)))).............................. ( -26.90) >DroSec_CAF1 36023 102 + 1 CUGGCUCUAAAGUUGAGCCGGACUAAAAAUAAAGUUUGGCCUUGUCAGCUUUAAGUUAAUCCACGAAAGAUGGGAAUAUAU--AUACAUAGAGUGAGAGAAAAG ((.(((((((((((((((((((((........))))))))....)))))))))......(((((....).)))).......--.......)))).))....... ( -31.10) >DroSim_CAF1 33973 102 + 1 CUGGCUCUAAAGUUGAGCCGGACUAAAAAUAAAGUUUGGCCUUGUCAGCUUUAAGUUAAUCCACGAAAGAUGGGAAUAUAU--AUACAUAGAGUGAGAGAAAAG ((.(((((((((((((((((((((........))))))))....)))))))))......(((((....).)))).......--.......)))).))....... ( -31.10) >DroYak_CAF1 35102 78 + 1 CUGGCUCUACAGUUAAGCCGGACUAAAAAUAAAGUUUGGCCUUGUCAGCUUUAAGUUUAUCCAGGGAAGAGAGGAACU-------------------------- ((..((((.(......((((((((........))))))))((((..(((.....)))....))))).)))).))....-------------------------- ( -16.60) >consensus CUGGCUCUAAAGUUGAGCCGGACUAAAAAUAAAGUUUGGCCUUGUCAGCUUUAAGUUAAUCCACGAAAGAUGGGAAUAUAU__AUACAUAGAGUGAGAGAAAAG .(((((.(((((((((((((((((........))))))))....)))))))))))))).(((.(....)...)))............................. (-20.85 = -21.85 + 1.00)

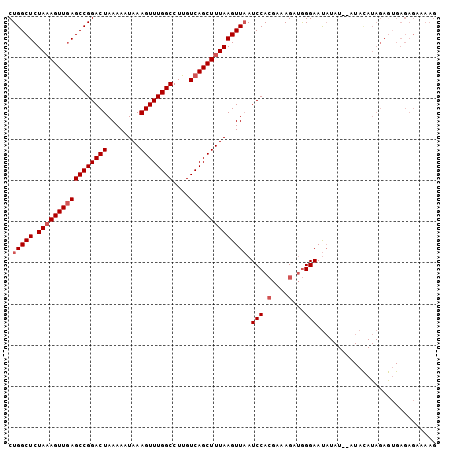
| Location | 16,580,767 – 16,580,871 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.04 |
| Mean single sequence MFE | -16.12 |
| Consensus MFE | -16.14 |
| Energy contribution | -16.20 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.09 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
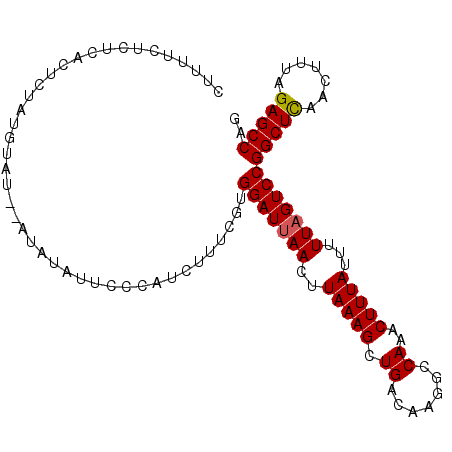
>2L_DroMel_CAF1 16580767 104 - 22407834 CUUUUCUCUCCUUCUAUAUAUACAUAUAUUCCCAUCUUUCGUGGAUUAACUUAAAGCUGACAAGGCCAAACUUUAUUUUUAGUCCGGCUCAACUUUAGAGCCAG ..........................................(((((((..(((((.((.......))..)))))...)))))))(((((.......))))).. ( -17.20) >DroSec_CAF1 36023 102 - 1 CUUUUCUCUCACUCUAUGUAU--AUAUAUUCCCAUCUUUCGUGGAUUAACUUAAAGCUGACAAGGCCAAACUUUAUUUUUAGUCCGGCUCAACUUUAGAGCCAG .....................--...................(((((((..(((((.((.......))..)))))...)))))))(((((.......))))).. ( -17.20) >DroSim_CAF1 33973 102 - 1 CUUUUCUCUCACUCUAUGUAU--AUAUAUUCCCAUCUUUCGUGGAUUAACUUAAAGCUGACAAGGCCAAACUUUAUUUUUAGUCCGGCUCAACUUUAGAGCCAG .....................--...................(((((((..(((((.((.......))..)))))...)))))))(((((.......))))).. ( -17.20) >DroYak_CAF1 35102 78 - 1 --------------------------AGUUCCUCUCUUCCCUGGAUAAACUUAAAGCUGACAAGGCCAAACUUUAUUUUUAGUCCGGCUUAACUGUAGAGCCAG --------------------------................(((((((..(((((.((.......))..)))))..))).))))(((((.......))))).. ( -12.90) >consensus CUUUUCUCUCACUCUAUGUAU__AUAUAUUCCCAUCUUUCGUGGAUUAACUUAAAGCUGACAAGGCCAAACUUUAUUUUUAGUCCGGCUCAACUUUAGAGCCAG ..........................................(((((((..(((((.((.......))..)))))...)))))))(((((.......))))).. (-16.14 = -16.20 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:48 2006