| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,578,530 – 16,578,629 |
| Length | 99 |
| Max. P | 0.914866 |

| Location | 16,578,530 – 16,578,629 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.02 |
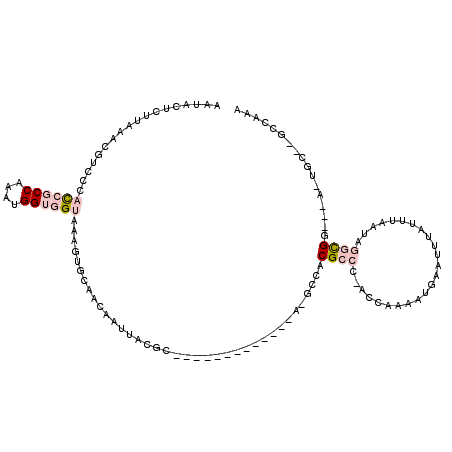
| Mean single sequence MFE | -31.57 |
| Consensus MFE | -5.77 |
| Energy contribution | -7.08 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.18 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
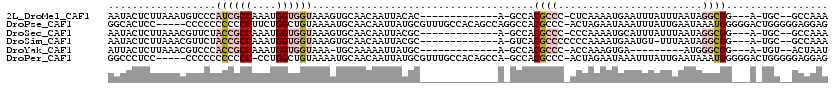
>2L_DroMel_CAF1 16578530 99 - 22407834 AAUACUCUUAAAUGUCCCAUCGCCAAAUGGUGGUAAAGUGCAACAAUUACAC-------------A-GCCACGCCC-CUCAAAAUGAAUUUAUUUAAUAGGCGG---A-UGC--GCCAAA .......(((((((....((((((....))))))...(((.........)))-------------.-.........-.............)))))))..((((.---.-..)--)))... ( -16.50) >DroPse_CAF1 42460 114 - 1 GGCACUCC-----CCCCCCCCCCCUUCUGGCUGUAAAAUGCAACAAUUAUGCGUUUGCCACAGCCAGGCCACGCCC-ACUAGAAUAAAUUUAUUGAAUAAAUGGGGGACUGGGGGAGGAG ....((((-----(((((.(((((.(((((((((.(((((((.......)))))))...))))))))).......(-(.((((.....)))).)).......)))))...))))).)))) ( -48.30) >DroSec_CAF1 33722 99 - 1 AAUACUCUUAAACGUUCUACCGCCAAAUGGUGGUAAAGUGCAACAAUUACGC-------------A-GCCACGCCC-CCCAAAAUGCAUUUAUUUAAUAGGCGG---A-UGC--GCCAAA .................(((((((....)))))))(((((((........((-------------.-.....))..-.......)))))))........((((.---.-..)--)))... ( -23.63) >DroSim_CAF1 31673 99 - 1 AAUACUCUUAAACGUUCUACCGCCAAAUGGUGGUAAAGUGCAACAAUUACGC-------------A-GUCACGCCCCCCCAAAAUGAAUGU-UUUAAUAGGCGG---A-UGC--GCCAAA .........(((((((((((((((....)))))))...(((.........))-------------)-..................))))))-)).....((((.---.-..)--)))... ( -25.00) >DroYak_CAF1 32761 89 - 1 AUUACUCUUAAACGUCCCACCGCCAAAUGGUGGUAAA-UGCAAAAAUUAUGC-------------A-GCCACGCCC-ACCAAAGUGA---------AUGGGCGG---A-UGU--ACUAAU ...........((((((.((((((....))))))...-((((.......)))-------------)-.....((((-(.........---------.)))))))---)-)))--...... ( -27.00) >DroPer_CAF1 41707 112 - 1 GGCCCUCC-----CCCCCCCCCCC-CCUGGCUGUAAAAUGCAACAAUUAUGCGUUUGCCACAGCCA-GCCACGCCC-ACUAGAAUAAAUUUAUUGAAUAAAUGGGGGACUGGGGGAGGAG ...(((((-----(((...(((((-.((((((((.(((((((.......)))))))...)))))))-).......(-(.((((.....)))).)).......)))))...)))))))).. ( -49.00) >consensus AAUACUCUUAAACGUCCCACCGCCAAAUGGUGGUAAAGUGCAACAAUUACGC_____________A_GCCACGCCC_ACCAAAAUGAAUUUAUUUAAUAGGCGG___A_UGC__GCCAAA ..................((((((....)))))).....................................((((........................))))................. ( -5.77 = -7.08 + 1.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:42 2006