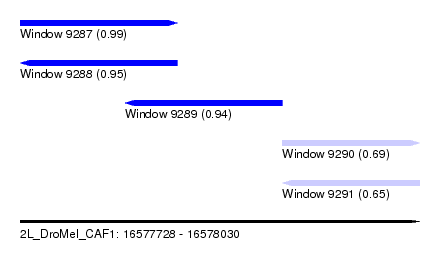
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,577,728 – 16,578,030 |
| Length | 302 |
| Max. P | 0.985921 |

| Location | 16,577,728 – 16,577,847 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.12 |
| Mean single sequence MFE | -24.41 |
| Consensus MFE | -22.90 |
| Energy contribution | -22.86 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16577728 119 + 22407834 CUUCUUACCUAAUUUUCCAGAUGCUAAUGGCUCAGUUUUUAACUCCACUUCAUUAGCUGGUGACCACGCCCAUUGGCGCAAAUAUUUUUUUUGGCCGCCUGCAUUUUAGAUCAUCGCCU ........((((.....(((..((((((((...(((.....))).....)))))))).((((....))))....(((.((((.......)))))))..)))....)))).......... ( -21.80) >DroSec_CAF1 32924 117 + 1 UUCCUCACCUAAUUUUCCAGAUGCUAAUGGCUCAGUUUUUAACUCCACUUCAUUAGCUGGUGGCCACGCCCCUUGGCUUAAAUAUUU--UUUGGCCGCCUGCAUUUUAGAUCAUCGCCU ...................(((((((((((...(((.....))))))........((.((((((((.(((....)))..(((....)--)))))))))).))...))))..)))).... ( -26.00) >DroSim_CAF1 30879 117 + 1 UUCCUCACCUAAUUUUUCAGAUGCUAAUGGCUCAGUUUUUAACUCCACUUCAUUAGCUGGUGGCCACGCCCCUUGGCUUAAAUAUUU--UUUGGCCGCCUGCAUUUUAGAUCAUCGCCU ...................(((((((((((...(((.....))))))........((.((((((((.(((....)))..(((....)--)))))))))).))...))))..)))).... ( -26.00) >DroEre_CAF1 31876 116 + 1 UUCCUCACCUAAUUUUCCCGAUGCUAAUGGCUCAGUUUUCAACUCCACUUCAUUAGCUGGUGGCCACGCC-CUUGGUUUAAAUAUUU--UUUGGCCGCCUGCAUUUUAGAUCAUCGCCU ..................((((((((((((...(((.....))))))........((.((((((((.(((-...)))..........--..)))))))).))...))))..)))))... ( -24.02) >DroYak_CAF1 32030 116 + 1 UUUCCCACCUAAUUUCCCAGAUGCUAAUGGCUCAGUCCUGAACUCCACUUCAUUAGCUGGUGGCCACGCC-CUUGGUUUAAAUAUUU--UUUGGCCGCCUGCAUUUUAGAUCAUCGCCU ...................(((((((((((.(((....)))...)))........((.((((((((.(((-...)))..........--..)))))))).))...))))..)))).... ( -24.22) >consensus UUCCUCACCUAAUUUUCCAGAUGCUAAUGGCUCAGUUUUUAACUCCACUUCAUUAGCUGGUGGCCACGCCCCUUGGCUUAAAUAUUU__UUUGGCCGCCUGCAUUUUAGAUCAUCGCCU ...................(((((((((((...(((.....))))))........((.((((((((.(((....)))..............)))))))).))...))))..)))).... (-22.90 = -22.86 + -0.04)

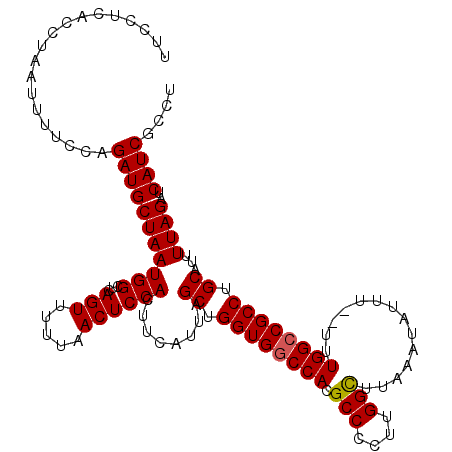
| Location | 16,577,728 – 16,577,847 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.12 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -27.42 |
| Energy contribution | -27.34 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.946837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16577728 119 - 22407834 AGGCGAUGAUCUAAAAUGCAGGCGGCCAAAAAAAAUAUUUGCGCCAAUGGGCGUGGUCACCAGCUAAUGAAGUGGAGUUAAAAACUGAGCCAUUAGCAUCUGGAAAAUUAGGUAAGAAG ........((((((......((.((((.(((......)))(((((....))))))))).)).(((((((..((..(((.....)))..)))))))))..........))))))...... ( -29.20) >DroSec_CAF1 32924 117 - 1 AGGCGAUGAUCUAAAAUGCAGGCGGCCAAA--AAAUAUUUAAGCCAAGGGGCGUGGCCACCAGCUAAUGAAGUGGAGUUAAAAACUGAGCCAUUAGCAUCUGGAAAAUUAGGUGAGGAA ......(.((((((......((.(((((..--..........(((....))).))))).)).(((((((..((..(((.....)))..)))))))))..........)))))).).... ( -30.52) >DroSim_CAF1 30879 117 - 1 AGGCGAUGAUCUAAAAUGCAGGCGGCCAAA--AAAUAUUUAAGCCAAGGGGCGUGGCCACCAGCUAAUGAAGUGGAGUUAAAAACUGAGCCAUUAGCAUCUGAAAAAUUAGGUGAGGAA ......(.((((((......((.(((((..--..........(((....))).))))).)).(((((((..((..(((.....)))..)))))))))..........)))))).).... ( -30.52) >DroEre_CAF1 31876 116 - 1 AGGCGAUGAUCUAAAAUGCAGGCGGCCAAA--AAAUAUUUAAACCAAG-GGCGUGGCCACCAGCUAAUGAAGUGGAGUUGAAAACUGAGCCAUUAGCAUCGGGAAAAUUAGGUGAGGAA ......(.((((((......((.(((((..--................-....))))).)).(((((((..((..(((.....)))..)))))))))..........)))))).).... ( -25.25) >DroYak_CAF1 32030 116 - 1 AGGCGAUGAUCUAAAAUGCAGGCGGCCAAA--AAAUAUUUAAACCAAG-GGCGUGGCCACCAGCUAAUGAAGUGGAGUUCAGGACUGAGCCAUUAGCAUCUGGGAAAUUAGGUGGGAAA ......(.((((((......((.(((((..--................-....))))).)).(((((((.......(((((....))))))))))))..........)))))).).... ( -26.26) >consensus AGGCGAUGAUCUAAAAUGCAGGCGGCCAAA__AAAUAUUUAAGCCAAGGGGCGUGGCCACCAGCUAAUGAAGUGGAGUUAAAAACUGAGCCAUUAGCAUCUGGAAAAUUAGGUGAGGAA ......(.((((((......((.(((((..............(((....))).))))).)).(((((((..((..((((...))))..)))))))))..........)))))).).... (-27.42 = -27.34 + -0.08)

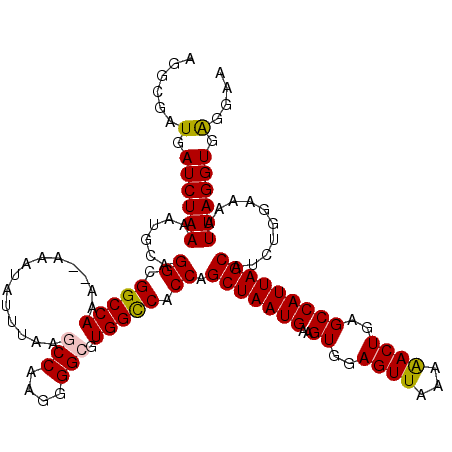
| Location | 16,577,807 – 16,577,926 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.28 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -22.30 |
| Energy contribution | -22.46 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16577807 119 - 22407834 CAAAAGGCAAAAAAGCAACGAUGAAAACGGGAGGAGGGGGUGGAGGUGGGGGCGUCCAGUGGAUAAUGGCCUUGUCAAAAGGCGAUGAUCUAAAAUGCAGGCGGCCAAAAAAAAUAUUU .....(((......(((.......................((((.((....)).)))).(((((.((.(((((.....))))).)).)))))...))).....)))............. ( -27.60) >DroSec_CAF1 33003 112 - 1 CAAAAGGCAAAAAAGCAACGAUGAAAACAGG-----GGGGCGGAGGUGGGCGCGUCCAGAGGAUGAUGGCCUUGUCAAAAGGCGAUGAUCUAAAAUGCAGGCGGCCAAA--AAAUAUUU .....(((......(((.............(-----((.(((........))).)))...((((.((.(((((.....))))).)).))))....))).....)))...--........ ( -31.30) >DroSim_CAF1 30958 112 - 1 CAAAAGGCAAAAAAGCAACGAUGAAAACAGG-----GGGGCGGAGGUGGGCGCGUCCAGAGGAUGAUGGCCUUGUCAAAAGGCGAUGAUCUAAAAUGCAGGCGGCCAAA--AAAUAUUU .....(((......(((.............(-----((.(((........))).)))...((((.((.(((((.....))))).)).))))....))).....)))...--........ ( -31.30) >DroEre_CAF1 31954 111 - 1 CAAAAGGCAAAAAAGCAACGAUGAAAAAAGG------GGGCGGAUGUGGGGGCAUCCGGAUGAUGAUGGGUUUGUCAAAAGGCGAUGAUCUAAAAUGCAGGCGGCCAAA--AAAUAUUU .....(((......(((..............------...(((((((....)))))))...(((.((.(.(((.....))).).)).))).....))).....)))...--........ ( -23.60) >DroYak_CAF1 32108 110 - 1 CAAAAGGCAAAAAAGCAACGAUGAAAAAAGG-------GGCGGAUGUGGGGGCGUCCGGAUGAUGAUGGGUUUGUCAAAAGGCGAUGAUCUAAAAUGCAGGCGGCCAAA--AAAUAUUU .....(((......(((..............-------..(((((((....)))))))...(((.((.(.(((.....))).).)).))).....))).....)))...--........ ( -23.20) >consensus CAAAAGGCAAAAAAGCAACGAUGAAAACAGG_____GGGGCGGAGGUGGGGGCGUCCAGAGGAUGAUGGCCUUGUCAAAAGGCGAUGAUCUAAAAUGCAGGCGGCCAAA__AAAUAUUU .....(((......(((........................(((.((....)).)))....(((.((.(((((.....))))).)).))).....))).....)))............. (-22.30 = -22.46 + 0.16)

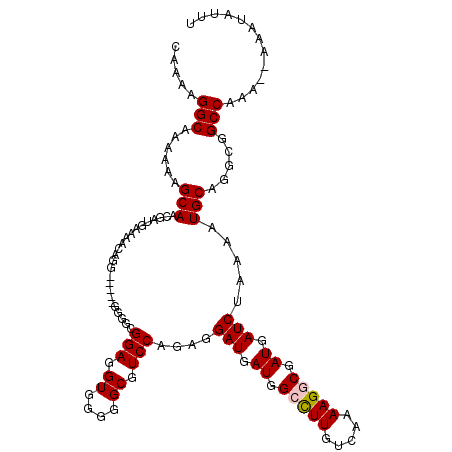
| Location | 16,577,926 – 16,578,030 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -16.69 |
| Energy contribution | -16.80 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16577926 104 + 22407834 GCGGUUUUCC--UUUUGU--UUGUUUAUGUGUGCGCUUGUGCUCUUUUUGGG-----------UGCUUUAAAUUAUAAAGUGUUAAUGGCGCUUUAAUAAACCGCUAAU-UUGUUGGUAA (((((((...--......--..................(..(((.....)))-----------..).........((((((((.....))))))))..)))))))....-.......... ( -22.80) >DroPse_CAF1 42216 100 + 1 GCGGUUUU-------------------UGUGUGUGUUUGUGCUCUUUUUGGGGGAAGGGGGCGGGCUUUAAAUUAUUAAGUGUUAAUGGCGCUUUAAUAAACCGCUAAU-UUGCUGGUAA (((((...-------------------.......((((((.(((((((....))))))).))))))......(((((((((((.....)))).))))))))))))....-.......... ( -27.50) >DroSim_CAF1 31070 106 + 1 GCGGUUUUCC--UUUUGUGUGUGUUUAUGUGUGCGCUUGUGCUCUUUUUGGG-----------GGCUUUAAAUUAUAAAGUGUUAAUGGCGCUUUAAUAAACCGCUAAU-UUGUUGGUAA (((((((...--....(((((..(....)..)))))....(((((.....))-----------))).........((((((((.....))))))))..)))))))....-.......... ( -26.20) >DroEre_CAF1 32065 106 + 1 GCGGUUUUCCUCUUUUGU--GUGUUUAUGUGUGCGCUUCUGCUCUUUUCGAG-----------GGCUUUAAAUUAUAAAGUGUUAAUGGCGCUUUAAUAAACCGCUAAU-UUGUUGGUAA (((((((.........((--(..(......)..)))....((((((...)))-----------))).........((((((((.....))))))))..)))))))....-.......... ( -25.70) >DroYak_CAF1 32218 103 + 1 GCGGUUUUCC--UUUUGU--GUGUUUAUG--UGCGCUUGUGCUCUUUUUGAG-----------AGCUUUAAAUUAUAAAGUGUUAAUGGCGCUUUAAUAAACCGCUAAUUUUGUUGGUAA (((((((...--....((--(..(....)--..)))....((((((...)))-----------))).........((((((((.....))))))))..)))))))............... ( -26.40) >DroPer_CAF1 41465 100 + 1 GCGGUUUU-------------------UGUGUGUGUUUGUGCUCUCUUUGGGGGAAGGGGGCGGGCUUUAAAUUAUUAAGUGUUAAUGGCGCUUUAAUAAACCGCUAAU-UUGCUGGUAA .((((...-------------------...(((.(((((((((((((((....)))))))))...............((((((.....))))))..)))))))))....-..)))).... ( -26.70) >consensus GCGGUUUUCC__UUUUGU__GUGUUUAUGUGUGCGCUUGUGCUCUUUUUGGG___________GGCUUUAAAUUAUAAAGUGUUAAUGGCGCUUUAAUAAACCGCUAAU_UUGUUGGUAA (((((((..................................(((.....))).......................((((((((.....))))))))..)))))))............... (-16.69 = -16.80 + 0.11)
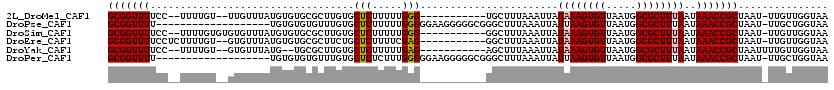
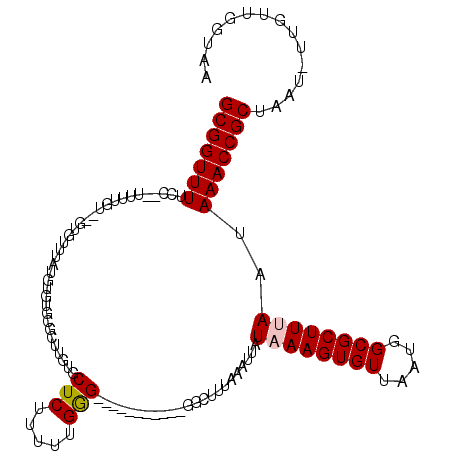
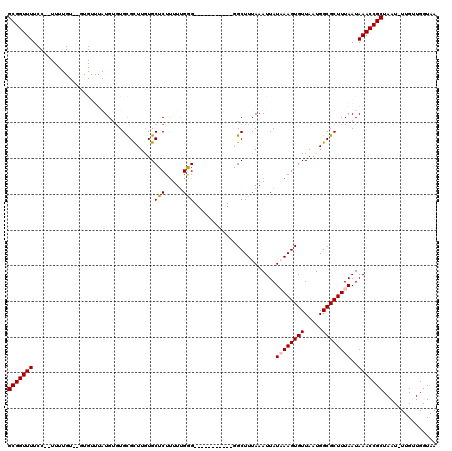
| Location | 16,577,926 – 16,578,030 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -14.30 |
| Consensus MFE | -9.17 |
| Energy contribution | -9.83 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16577926 104 - 22407834 UUACCAACAA-AUUAGCGGUUUAUUAAAGCGCCAUUAACACUUUAUAAUUUAAAGCA-----------CCCAAAAAGAGCACAAGCGCACACAUAAACAA--ACAAAA--GGAAAACCGC ..........-....(((((((......((((.........((((.....))))((.-----------..........))....))))............--.(....--)..))))))) ( -13.60) >DroPse_CAF1 42216 100 - 1 UUACCAGCAA-AUUAGCGGUUUAUUAAAGCGCCAUUAACACUUAAUAAUUUAAAGCCCGCCCCCUUCCCCCAAAAAGAGCACAAACACACACA-------------------AAAACCGC ..........-....(((((((......((...((((........)))).....))..((...(((........))).)).............-------------------.))))))) ( -9.00) >DroSim_CAF1 31070 106 - 1 UUACCAACAA-AUUAGCGGUUUAUUAAAGCGCCAUUAACACUUUAUAAUUUAAAGCC-----------CCCAAAAAGAGCACAAGCGCACACAUAAACACACACAAAA--GGAAAACCGC ..........-....(((((((......((((.........((((.....))))(((-----------........).))....)))).............(......--)..))))))) ( -13.80) >DroEre_CAF1 32065 106 - 1 UUACCAACAA-AUUAGCGGUUUAUUAAAGCGCCAUUAACACUUUAUAAUUUAAAGCC-----------CUCGAAAAGAGCAGAAGCGCACACAUAAACAC--ACAAAAGAGGAAAACCGC ..........-....(((((((......((((........(((((.....)))))..-----------(((.....))).....))))............--.(......)..))))))) ( -18.00) >DroYak_CAF1 32218 103 - 1 UUACCAACAAAAUUAGCGGUUUAUUAAAGCGCCAUUAACACUUUAUAAUUUAAAGCU-----------CUCAAAAAGAGCACAAGCGCA--CAUAAACAC--ACAAAA--GGAAAACCGC ...............(((((((......((((.........((((.....))))(((-----------((.....)))))....)))).--........(--......--)..))))))) ( -20.10) >DroPer_CAF1 41465 100 - 1 UUACCAGCAA-AUUAGCGGUUUAUUAAAGCGCCAUUAACACUUAAUAAUUUAAAGCCCGCCCCCUUCCCCCAAAGAGAGCACAAACACACACA-------------------AAAACCGC ..........-....(((((((......((...((((........)))).....))..((.(.(((......))).).)).............-------------------.))))))) ( -11.30) >consensus UUACCAACAA_AUUAGCGGUUUAUUAAAGCGCCAUUAACACUUUAUAAUUUAAAGCC___________CCCAAAAAGAGCACAAGCGCACACAUAAACAC__ACAAAA__GGAAAACCGC ...............(((((((..................(((((.....))))).......................((......)).........................))))))) ( -9.17 = -9.83 + 0.67)

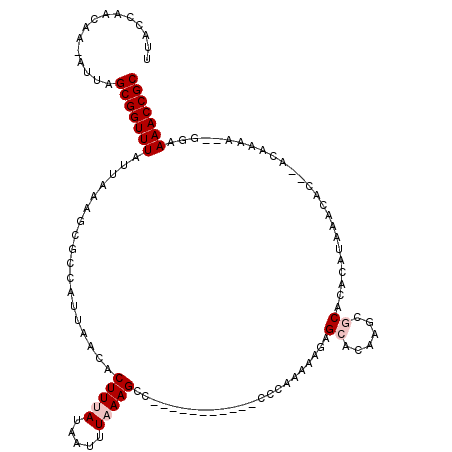
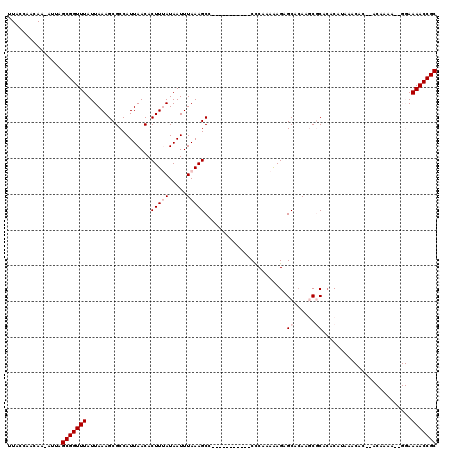
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:41 2006