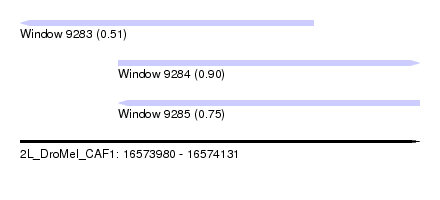
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,573,980 – 16,574,131 |
| Length | 151 |
| Max. P | 0.897565 |

| Location | 16,573,980 – 16,574,091 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.44 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -16.17 |
| Energy contribution | -16.17 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
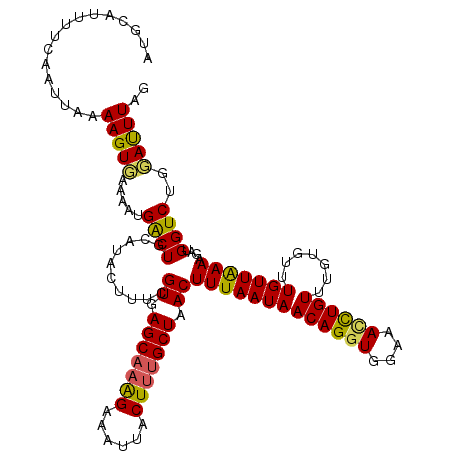
>2L_DroMel_CAF1 16573980 111 - 22407834 GUGAGCAAAGAAAUUACUUUGCUAACUUUAAUAACAGGUGGAAACCUGUU------UGUUAAAAUAUGGUCUGUAUUUAGU---UUUUCAUUUUCAAGAUUGCAACUUGAGUAUAAAUGA ((.(((((((......))))))).))(((((((((((((....))))).)------)))))))..................---...(((((((((((.......)))))....)))))) ( -27.20) >DroSec_CAF1 29113 116 - 1 GUGAGCAAAGAAAUUACUUGGCUAACUUUAAUAACAGGUGGAAACCUGUUUGUGUUUGUUAAAAUAUGGUCUGUAUUUAGC---UUUUAAUUUUCAAGAUUGGAACUUGAAAA-AAAAGA ..((((....((((.((..((((.........(((((((....))))))).((((((....)))))))))).)))))).))---))....((((((((.......))))))))-...... ( -24.70) >DroEre_CAF1 28180 116 - 1 GUGAGCAAAGGAAUUACUUUGCUAACUUUAAUAACAGGUGGACAGCUGUUUUUGUUUGUUGAAAGAUGGUCUAGAUUUAGC---UUUUC-AUUUCAAGAUUGGAGUACGAGUAUAUAAUA ...(((((((......))))))).((((((((...(((((((.(((((..((((.(..((....))..)..))))..))))---).)))-))))....)))))))).............. ( -30.10) >DroYak_CAF1 28176 119 - 1 GUGAGCAAGGAAAUUACUAUGCUGACUUUAAUAACAGGUGGAAAUUUGUGUGUGUUUGUUGAAAGAUGGUCUGGACUUAGCAAAUUGGA-CUUUGUAGAUUGGGAUUCGAGUACAUAAUA ((.((((.((......)).)))).)).......((((((....))))))(((((((((.........(((((.((.((.(((((.....-.))))).)))).)))))))))))))).... ( -22.20) >consensus GUGAGCAAAGAAAUUACUUUGCUAACUUUAAUAACAGGUGGAAACCUGUUUGUGUUUGUUAAAAGAUGGUCUGGAUUUAGC___UUUUA_UUUUCAAGAUUGGAACUCGAGUAUAAAAGA ((.(((((((......))))))).))(((((((((((((....)))))).......)))))))....(((((........................)))))................... (-16.17 = -16.17 + -0.00)

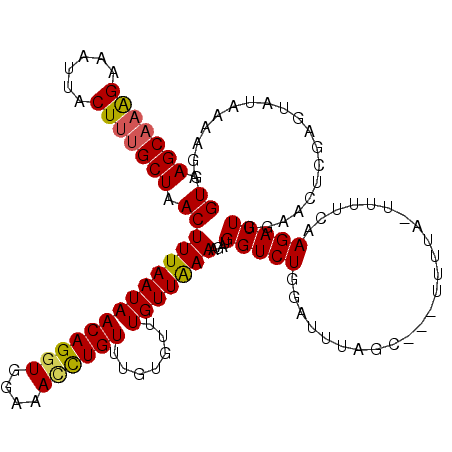

| Location | 16,574,017 – 16,574,131 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.47 |
| Mean single sequence MFE | -22.39 |
| Consensus MFE | -16.14 |
| Energy contribution | -18.70 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16574017 114 + 22407834 CUAAAUACAGACCAUAUUUUAACA------AACAGGUUUCCACCUGUUAUUAAAGUUAGCAAAGUAAUUUCUUUGCUCACGAAAGUAUGCAGUCAUUUUCACUUUUAAUUGAAAAUGCAU ............((((((((....------(((((((....)))))))......((.(((((((......))))))).)).))))))))..(.((((((((........))))))))).. ( -29.30) >DroSec_CAF1 29149 120 + 1 CUAAAUACAGACCAUAUUUUAACAAACACAAACAGGUUUCCACCUGUUAUUAAAGUUAGCCAAGUAAUUUCUUUGCUCACGAAAGUAUGCAGUCAUUUUCACUUUUAAUUGAAAAUGCAU ............((((((((..........(((((((....)))))))......((.(((.(((......))).))).)).))))))))..(.((((((((........))))))))).. ( -25.20) >DroEre_CAF1 28216 120 + 1 CUAAAUCUAGACCAUCUUUCAACAAACAAAAACAGCUGUCCACCUGUUAUUAAAGUUAGCAAAGUAAUUCCUUUGCUCACGAAAGUAUGCAGCCCUUUUGACUUUUAAUUGAAAAUGCAU ............((((((((..........(((((........)))))......((.(((((((......))))))).))))))).)))..((..(((..(.......)..)))..)).. ( -20.90) >DroYak_CAF1 28215 120 + 1 CUAAGUCCAGACCAUCUUUCAACAAACACACACAAAUUUCCACCUGUUAUUAAAGUCAGCAUAGUAAUUUCCUUGCUCACGAAAGUAUGCAGCCAUUUUCACUUUUAAUUGGAAAAGCAU ............((((((((((((....................))))......((.((((..(......)..)))).))))))).)))..((..((((((........)))))).)).. ( -14.15) >consensus CUAAAUACAGACCAUAUUUCAACAAACACAAACAGGUUUCCACCUGUUAUUAAAGUUAGCAAAGUAAUUUCUUUGCUCACGAAAGUAUGCAGCCAUUUUCACUUUUAAUUGAAAAUGCAU ............((((((((..........(((((((....)))))))......((.(((((((......))))))).))))))).)))..(.((((((((........))))))))).. (-16.14 = -18.70 + 2.56)

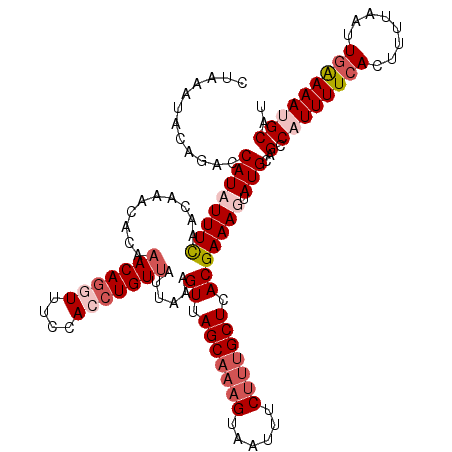
| Location | 16,574,017 – 16,574,131 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.47 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -20.75 |
| Energy contribution | -20.31 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16574017 114 - 22407834 AUGCAUUUUCAAUUAAAAGUGAAAAUGACUGCAUACUUUCGUGAGCAAAGAAAUUACUUUGCUAACUUUAAUAACAGGUGGAAACCUGUU------UGUUAAAAUAUGGUCUGUAUUUAG ...((((((((........)))))))).(((.((((..((((((((((((......)))))))...(((((((((((((....))))).)------))))))).)))))...)))).))) ( -33.50) >DroSec_CAF1 29149 120 - 1 AUGCAUUUUCAAUUAAAAGUGAAAAUGACUGCAUACUUUCGUGAGCAAAGAAAUUACUUGGCUAACUUUAAUAACAGGUGGAAACCUGUUUGUGUUUGUUAAAAUAUGGUCUGUAUUUAG ...((((((((........)))))))).(((.((((..(((((.((.(((......))).))((((......(((((((....))))))).......))))...)))))...)))).))) ( -27.32) >DroEre_CAF1 28216 120 - 1 AUGCAUUUUCAAUUAAAAGUCAAAAGGGCUGCAUACUUUCGUGAGCAAAGGAAUUACUUUGCUAACUUUAAUAACAGGUGGACAGCUGUUUUUGUUUGUUGAAAGAUGGUCUAGAUUUAG ...((((((((((...(((..((((.(((((..(((((..((.(((((((......))))))).)).........)))))..))))).))))..))))))).))))))............ ( -27.30) >DroYak_CAF1 28215 120 - 1 AUGCUUUUCCAAUUAAAAGUGAAAAUGGCUGCAUACUUUCGUGAGCAAGGAAAUUACUAUGCUGACUUUAAUAACAGGUGGAAAUUUGUGUGUGUUUGUUGAAAGAUGGUCUGGACUUAG .......((((.......((....))((((((((((....((.((((.((......)).)))).)).......((((((....))))))))))))..(((....)))))))))))..... ( -24.40) >consensus AUGCAUUUUCAAUUAAAAGUGAAAAUGACUGCAUACUUUCGUGAGCAAAGAAAUUACUUUGCUAACUUUAAUAACAGGUGGAAACCUGUUUGUGUUUGUUAAAAGAUGGUCUGGAUUUAG ................(((((.....((((..........((.(((((((......))))))).))(((((((((((((....)))))).......)))))))....))))..))))).. (-20.75 = -20.31 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:35 2006