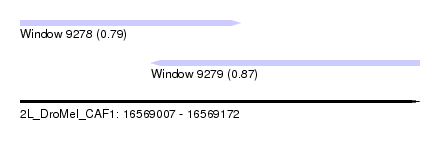
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,569,007 – 16,569,172 |
| Length | 165 |
| Max. P | 0.868952 |

| Location | 16,569,007 – 16,569,098 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.31 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -14.57 |
| Energy contribution | -17.32 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16569007 91 + 22407834 AGCAGCAACACCCAAGUGCAGCAGCAACAUCAA-CUAUAAUAAGUAC-------CAAGGGCUGGAUGAAGUUGAGAGGCUGCGAAUGGCGCUUGG---UGUU ......((((((.((((((.(((((....((((-((.((...(((.(-------....))))...)).))))))...))))).....))))))))---)))) ( -33.40) >DroSec_CAF1 23985 91 + 1 AGCAGCAACACCCAAGUGCAGCAGCAACAUCAA-CUAUAAUAAGUAC-------CAAGGGCUGGAUGAAGUUGAGAGGCUGCGAAUGGCGCUUGG---UGUU ......((((((.((((((.(((((....((((-((.((...(((.(-------....))))...)).))))))...))))).....))))))))---)))) ( -33.40) >DroSim_CAF1 22071 91 + 1 AGCAGCAACACCCAAGUGCAGCAGCAACAUCAA-CUAUAAUAAGUAC-------CAAGGGCUGGAUGAAGUUGAGAGGCUGCGAAUGGCGCUUGG---UGUU ......((((((.((((((.(((((....((((-((.((...(((.(-------....))))...)).))))))...))))).....))))))))---)))) ( -33.40) >DroWil_CAF1 25722 100 + 1 AGCAGCAACAGACAACAACAACAACAGCAGCAAAGUAGUAUAAGCAGAAGAAUACAAAGUCGAGAGGGAACUGAG--ACUGCAACUGGCGCUUGGUGGUGUU ..........((((.((.((((..(((..((......((((..........))))..((((...((....))..)--)))))..)))..).))).)).)))) ( -16.40) >consensus AGCAGCAACACCCAAGUGCAGCAGCAACAUCAA_CUAUAAUAAGUAC_______CAAGGGCUGGAUGAAGUUGAGAGGCUGCGAAUGGCGCUUGG___UGUU ...((((....((((((((.(((((....((((.........(((..............)))........))))...))))).....))))))))...)))) (-14.57 = -17.32 + 2.75)


| Location | 16,569,061 – 16,569,172 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.43 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -18.11 |
| Energy contribution | -18.87 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16569061 111 - 22407834 GGCGAA-----CCGAUGCACAACAGCAACAUGCAACAA-CAUCGCGAAUGGCAAGUGGUAGUCAUAACAACAAUGCCGUCAACACCAAGCGCCAUUCGCAGCCUCUCAACUUCAUCC (((...-----..((((.......((.....)).....-))))(((((((((...((((.((...................))))))...))))))))).))).............. ( -29.21) >DroPse_CAF1 30907 103 - 1 AUCGUG-----CUGGUGCUG---------GUGCUGUAGCUGUAGCUGGUGGCAAGUGGUAGUCAUAAUAACAAUGCCGUCAACACCAAGCGCCAGUCGCAGCCUCUCAGCUUCUUUC ...(((-----(((((((((---------(((....(((....)))(..((((..((.............)).))))..)..)))).)))))))).)))(((......)))...... ( -35.12) >DroSim_CAF1 22125 111 - 1 GGCGAA-----CCGAUGCACAACAGCAACAUGCAACAA-CAUCGCGGAUGGCAAGUGGUAGUCAUAACAACAAUGCCGUCAACACCAAGCGCCAUUCGCAGCCUCUCAACUUCAUCC (((...-----..((((.......((.....)).....-))))(((((((((...((((.((...................))))))...))))))))).))).............. ( -28.71) >DroEre_CAF1 23379 116 - 1 GGCAAACCGAGCCGAGGCACACCAGCAACGUGCAACAA-CAUCGCGGAUGGCAAGUGGUAGUCAUAACAACAGUGCCGUCAACACCAAGCGCCAUUCGCAGCCUCUCAACUUCAUCC (((.......)))(((((......((.....)).....-....(((((((((...((((.((....((....)).......))))))...))))))))).)))))............ ( -36.20) >DroYak_CAF1 23384 116 - 1 GGCCAACCGAACCAAUGCACAACAGCAACAUGCAACAA-CAUCGCGGAUGGCAAGUGGUAGUCAUAACAACAAUGCCGCCAACACCAAGCGCCAUUCGCAGCCUCUCAACUUCAUCC (((............(((......)))...........-....(((((((((....((((.............))))((.........))))))))))).))).............. ( -27.22) >DroPer_CAF1 32474 91 - 1 AUCGUG-----CUGGUGC---------------------UGUAGCUGGUGGCAAGUGGUAGUCAUAAUAACAAUGCCGUCAACACCAAGCGCCAGUCGCAGCCUCUCAGCUUCUUUC .....(-----((((.((---------------------((..(((((((.(...((((.((...................)))))).))))))))..))))..).))))....... ( -28.51) >consensus GGCGAA_____CCGAUGCACAACAGCAACAUGCAACAA_CAUCGCGGAUGGCAAGUGGUAGUCAUAACAACAAUGCCGUCAACACCAAGCGCCAUUCGCAGCCUCUCAACUUCAUCC .............((.((......((.....))..........(((((((((...((((.(((((.......)))......))))))...))))))))).)).))............ (-18.11 = -18.87 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:29 2006