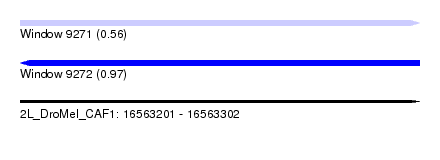
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,563,201 – 16,563,302 |
| Length | 101 |
| Max. P | 0.969939 |

| Location | 16,563,201 – 16,563,302 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.80 |
| Mean single sequence MFE | -24.88 |
| Consensus MFE | -8.89 |
| Energy contribution | -8.89 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16563201 101 + 22407834 CGUGCA--ACGUAACAAAUUU-ACCCGUAGACAGAUUGCACAUGUGUACCGUUUUGUAAUGUUAUAC-AAGUA-------ACACAUGCACACACUUUUGGCUACACCGCACA-------- .((((.--..((((.....))-))..((((.((((.((..(((((((.....((((((......)))-)))..-------)))))))..))....)))).))))...)))).-------- ( -24.90) >DroSim_CAF1 16140 101 + 1 CGUGCA--ACGUAACAAAUUU-ACCCGUAGACAGAUUGCACAUGUGUACCGUUUUGUAAUGUUAUAC-AAGUA-------ACACAUGCACACACUUUUGGCUACACCGCACA-------- .((((.--..((((.....))-))..((((.((((.((..(((((((.....((((((......)))-)))..-------)))))))..))....)))).))))...)))).-------- ( -24.90) >DroEre_CAF1 17089 116 + 1 CGUGCA--ACGUAACAAAUUU-ACCCGUAGACAGAUUGCACAUGUGUACCGUUUUGUAAUGUUAUAC-AAGUACGGGUAUACACAUGCACACACUUUUGUCUACACCGCACACAAACACA .((((.--..((((.....))-))..(((((((((.((..((((((((((((((((((......)))-))).))))...))))))))..))....)))))))))...))))......... ( -37.30) >DroWil_CAF1 20276 87 + 1 CGUGCAAAACGUAACAAAUUU-G-CAGCAGACAGAUUGCACAUGUGUAACGUUUUGUAAUGCUAUACCAUGUA-------ACACAGCGUCGGGUU-----------A------------- ..((((((((((.(((.((.(-(-((((.....).))))).)).))).))))))))))........((((((.-------.....)))).))...-----------.------------- ( -22.50) >DroYak_CAF1 17631 101 + 1 CGUGCA--ACGUAACAAAUUU-ACCCGUAGACAGAUUGCACAUGUGUACCGUUUUGUAAUGUUAUAC-AAGUA-------ACACAUGCUCACGCUUUUAGCUACACCGCACA-------- .((((.--..((((.....))-))..((((.(.((..((.(((((((.....((((((......)))-)))..-------))))))).....))..)).)))))...)))).-------- ( -23.30) >DroPer_CAF1 25591 86 + 1 CGUGCA--ACGUAACAAAUUUAACCCGUGGACAGAUUGCACAUGUGUACCGUUUUGUAAUGUUAUAC-AAGUA-------GCACACACACACACA-----------C------------- .((((.--((................(((.(.....).))).((((((.((((....)))).)))))-).)).-------))))...........-----------.------------- ( -16.40) >consensus CGUGCA__ACGUAACAAAUUU_ACCCGUAGACAGAUUGCACAUGUGUACCGUUUUGUAAUGUUAUAC_AAGUA_______ACACAUGCACACACUUUUGGCUACACCGCACA________ .(((((....((..................))....)))))..(((((.((((....)))).)))))..................................................... ( -8.89 = -8.89 + 0.00)


| Location | 16,563,201 – 16,563,302 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.80 |
| Mean single sequence MFE | -32.43 |
| Consensus MFE | -14.05 |
| Energy contribution | -15.05 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16563201 101 - 22407834 --------UGUGCGGUGUAGCCAAAAGUGUGUGCAUGUGU-------UACUU-GUAUAACAUUACAAAACGGUACACAUGUGCAAUCUGUCUACGGGU-AAAUUUGUUACGU--UGCACG --------.((((..((((((........((..(((((((-------.((((-(((......))))....))))))))))..))(((((....)))))-......)))))).--.)))). ( -36.20) >DroSim_CAF1 16140 101 - 1 --------UGUGCGGUGUAGCCAAAAGUGUGUGCAUGUGU-------UACUU-GUAUAACAUUACAAAACGGUACACAUGUGCAAUCUGUCUACGGGU-AAAUUUGUUACGU--UGCACG --------.((((..((((((........((..(((((((-------.((((-(((......))))....))))))))))..))(((((....)))))-......)))))).--.)))). ( -36.20) >DroEre_CAF1 17089 116 - 1 UGUGUUUGUGUGCGGUGUAGACAAAAGUGUGUGCAUGUGUAUACCCGUACUU-GUAUAACAUUACAAAACGGUACACAUGUGCAAUCUGUCUACGGGU-AAAUUUGUUACGU--UGCACG .........((((..(((.((((((....((..((((((((...((((..((-(((......))))).))))))))))))..))(((((....)))))-...))))))))).--.)))). ( -41.70) >DroWil_CAF1 20276 87 - 1 -------------U-----------AACCCGACGCUGUGU-------UACAUGGUAUAGCAUUACAAAACGUUACACAUGUGCAAUCUGUCUGCUG-C-AAAUUUGUUACGUUUUGCACG -------------.-----------........((((((.-------.......))))))....((((((((.(((.((.((((..........))-)-).)).))).)))))))).... ( -20.00) >DroYak_CAF1 17631 101 - 1 --------UGUGCGGUGUAGCUAAAAGCGUGAGCAUGUGU-------UACUU-GUAUAACAUUACAAAACGGUACACAUGUGCAAUCUGUCUACGGGU-AAAUUUGUUACGU--UGCACG --------.((((..((((((........((.((((((((-------.((((-(((......))))....))))))))))).))(((((....)))))-......)))))).--.)))). ( -33.10) >DroPer_CAF1 25591 86 - 1 -------------G-----------UGUGUGUGUGUGUGC-------UACUU-GUAUAACAUUACAAAACGGUACACAUGUGCAAUCUGUCCACGGGUUAAAUUUGUUACGU--UGCACG -------------(-----------((((..(((((((((-------(..((-(((......)))))...))))))))))..)((((((....)))))).............--.)))). ( -27.40) >consensus ________UGUGCGGUGUAGCCAAAAGUGUGUGCAUGUGU_______UACUU_GUAUAACAUUACAAAACGGUACACAUGUGCAAUCUGUCUACGGGU_AAAUUUGUUACGU__UGCACG ..........................((((((((..(((........)))...(((......)))......))))))))(((((...(((..((((((...)))))).)))...))))). (-14.05 = -15.05 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:22 2006