
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,556,448 – 16,556,648 |
| Length | 200 |
| Max. P | 0.998642 |

| Location | 16,556,448 – 16,556,568 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -39.78 |
| Consensus MFE | -35.58 |
| Energy contribution | -35.46 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16556448 120 + 22407834 GAAACCGUUGGGCGCUGCCUUUGUGUGCGGAUUGUUUGCUUUUUGGGCCUGGCCAAAGGUAGCGAAAGCAUUGGUUAAGUUCUUGGCUUGUUUGCCCGGCUUAUUUGCCAAAUAAUCUGU .........((((((((((((((.((.((((..(....)..)))).)).....))))))))))..(((((..((((((....)))))))))))))))(((......)))........... ( -39.00) >DroSec_CAF1 10974 120 + 1 GAAACCGUUGGGCGCUGCCUUUGUGUGCGGAUUGUUUGCUUUUUGGGCCUGGCCAAAGGUAGCGAAAGCAUUGGUUAAGUUCUUGGCUUGUUUGCCCGGCUUAUUUGCCAAAUAAUCUGU .........((((((((((((((.((.((((..(....)..)))).)).....))))))))))..(((((..((((((....)))))))))))))))(((......)))........... ( -39.00) >DroSim_CAF1 10923 120 + 1 GAAACCGUUGGGCGCUGCCUUUGUGUGCGGAUUGUUUGCUUUUUGGGCCUGGCCAAAGGUAGCGAAAGCAUUGGUUAAGUUCUUGGCUUGUUUGCCCGGCUUAUUUGCCAAAUAAUCUGU .........((((((((((((((.((.((((..(....)..)))).)).....))))))))))..(((((..((((((....)))))))))))))))(((......)))........... ( -39.00) >DroEre_CAF1 10816 120 + 1 GAAACCGUUGGGCGCUCCCUUUGUGUGCGAAUUGUUUGCUUUUUGGGCCUGUCCAAAGGUAGCGAAAGCAUUGGUUAAGUUCUUGGCUUGUUUGCCCAGCUUAUUUGCCAAAUAAUCUGU ...(((.(((((((..(((.......((((.....)))).....)))..))))))).))).((....)).(((((((((((...(((......))).))))))...)))))......... ( -37.50) >DroYak_CAF1 10783 120 + 1 GAAACCAUUGGGCGCUCCCUUUGUGUGCGGAUUGUUUGCUUUUUGGGCCUGGCCAAAGGUAGCGAAAGCAUUGGUUAAGUUCUUGGCUUGUUUGCCAGGCUUAUUUGCCAAAUAAUCUGU ...((((..(((....)))..)).))(((((((((((((....((((((((((.(((.(..((....))...((((((....))))))).)))))))))))))...).)))))))))))) ( -44.40) >consensus GAAACCGUUGGGCGCUGCCUUUGUGUGCGGAUUGUUUGCUUUUUGGGCCUGGCCAAAGGUAGCGAAAGCAUUGGUUAAGUUCUUGGCUUGUUUGCCCGGCUUAUUUGCCAAAUAAUCUGU ...((((..((......))..)).))(((((((((.(((((((...((((......))))...)))))))(((((((((((...(((......))).))))))...)))))))))))))) (-35.58 = -35.46 + -0.12)


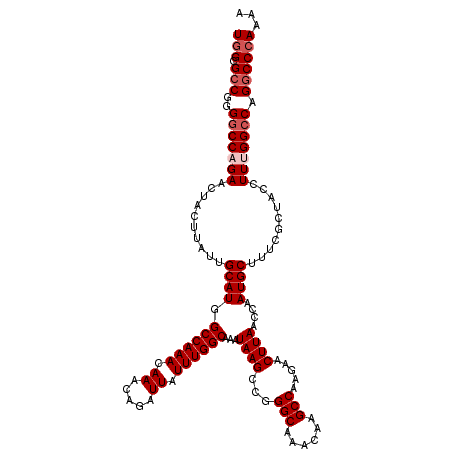
| Location | 16,556,488 – 16,556,608 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -40.92 |
| Consensus MFE | -38.94 |
| Energy contribution | -38.98 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.998642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16556488 120 + 22407834 UUUUGGGCCUGGCCAAAGGUAGCGAAAGCAUUGGUUAAGUUCUUGGCUUGUUUGCCCGGCUUAUUUGCCAAAUAAUCUGUUUGUUUGGCCAUGCAAUAAGUAGUUCUGGCCCCGGCGCCA ...((((((.(((((..((((((....))...((((((....))))))....))))..(((((((.(((((((((.....))))))))).....))))))).....)))))..))).))) ( -42.10) >DroSec_CAF1 11014 120 + 1 UUUUGGGCCUGGCCAAAGGUAGCGAAAGCAUUGGUUAAGUUCUUGGCUUGUUUGCCCGGCUUAUUUGCCAAAUAAUCUGUUUGUUUGGCCAUGCAAUAAGUAGUUCGGGCCCCGGCGCCA ....(((((((..(...((((((....))...((((((....))))))....))))..(((((((.(((((((((.....))))))))).....))))))).)..)))))))........ ( -41.50) >DroSim_CAF1 10963 120 + 1 UUUUGGGCCUGGCCAAAGGUAGCGAAAGCAUUGGUUAAGUUCUUGGCUUGUUUGCCCGGCUUAUUUGCCAAAUAAUCUGUUUGUUUGGCCAUGCAAUAAGUAGUUCGGGCCCCGGCGCCA ....(((((((..(...((((((....))...((((((....))))))....))))..(((((((.(((((((((.....))))))))).....))))))).)..)))))))........ ( -41.50) >DroEre_CAF1 10856 120 + 1 UUUUGGGCCUGUCCAAAGGUAGCGAAAGCAUUGGUUAAGUUCUUGGCUUGUUUGCCCAGCUUAUUUGCCAAAUAAUCUGUUUGUUUGGCCAUGCAAUAAGUAGUUCUGGCCCCGUCGCCA ....(((((.(..(...((((((....))...((((((....))))))....))))..(((((((.(((((((((.....))))))))).....))))))).)..).)))))........ ( -36.70) >DroYak_CAF1 10823 120 + 1 UUUUGGGCCUGGCCAAAGGUAGCGAAAGCAUUGGUUAAGUUCUUGGCUUGUUUGCCAGGCUUAUUUGCCAAAUAAUCUGUUUGUUUGGCCAUGCAAUAAGUAGUUCUGGCCCCGGCGCUA ....(((((((((((((.(..((....)).(((((.((((.((((((......))))))...)))))))))..........).))))))))(((.....))).....)))))........ ( -42.80) >consensus UUUUGGGCCUGGCCAAAGGUAGCGAAAGCAUUGGUUAAGUUCUUGGCUUGUUUGCCCGGCUUAUUUGCCAAAUAAUCUGUUUGUUUGGCCAUGCAAUAAGUAGUUCUGGCCCCGGCGCCA ....(((((((((((((.(..((....)).(((((((((((...(((......))).))))))...)))))..........).))))))))(((.....))).....)))))........ (-38.94 = -38.98 + 0.04)


| Location | 16,556,488 – 16,556,608 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -33.22 |
| Consensus MFE | -30.39 |
| Energy contribution | -31.39 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16556488 120 - 22407834 UGGCGCCGGGGCCAGAACUACUUAUUGCAUGGCCAAACAAACAGAUUAUUUGGCAAAUAAGCCGGGCAAACAAGCCAAGAACUUAACCAAUGCUUUCGCUACCUUUGGCCAGGCCCAAAA (((.(((..(((((((..........((((.((((((.((.....)).))))))...((((...(((......))).....))))....))))..........))))))).))))))... ( -36.45) >DroSec_CAF1 11014 120 - 1 UGGCGCCGGGGCCCGAACUACUUAUUGCAUGGCCAAACAAACAGAUUAUUUGGCAAAUAAGCCGGGCAAACAAGCCAAGAACUUAACCAAUGCUUUCGCUACCUUUGGCCAGGCCCAAAA (((.(((.(((((((.....((((((.....((((((.((.....)).)))))).)))))).)))))...................((((.((....)).....)))))).))))))... ( -33.20) >DroSim_CAF1 10963 120 - 1 UGGCGCCGGGGCCCGAACUACUUAUUGCAUGGCCAAACAAACAGAUUAUUUGGCAAAUAAGCCGGGCAAACAAGCCAAGAACUUAACCAAUGCUUUCGCUACCUUUGGCCAGGCCCAAAA (((.(((.(((((((.....((((((.....((((((.((.....)).)))))).)))))).)))))...................((((.((....)).....)))))).))))))... ( -33.20) >DroEre_CAF1 10856 120 - 1 UGGCGACGGGGCCAGAACUACUUAUUGCAUGGCCAAACAAACAGAUUAUUUGGCAAAUAAGCUGGGCAAACAAGCCAAGAACUUAACCAAUGCUUUCGCUACCUUUGGACAGGCCCAAAA ........(((((.......((((((.....((((((.((.....)).)))))).))))))((.(((......))).)).......((((.((....)).....))))...))))).... ( -30.00) >DroYak_CAF1 10823 120 - 1 UAGCGCCGGGGCCAGAACUACUUAUUGCAUGGCCAAACAAACAGAUUAUUUGGCAAAUAAGCCUGGCAAACAAGCCAAGAACUUAACCAAUGCUUUCGCUACCUUUGGCCAGGCCCAAAA ..(.(((..(((((((..........((((.((((((.((.....)).))))))...((((..((((......))))....))))....))))..........))))))).))).).... ( -33.25) >consensus UGGCGCCGGGGCCAGAACUACUUAUUGCAUGGCCAAACAAACAGAUUAUUUGGCAAAUAAGCCGGGCAAACAAGCCAAGAACUUAACCAAUGCUUUCGCUACCUUUGGCCAGGCCCAAAA (((.(((..(((((((..........((((.((((((.((.....)).))))))...((((...(((......))).....))))....))))..........))))))).))))))... (-30.39 = -31.39 + 1.00)


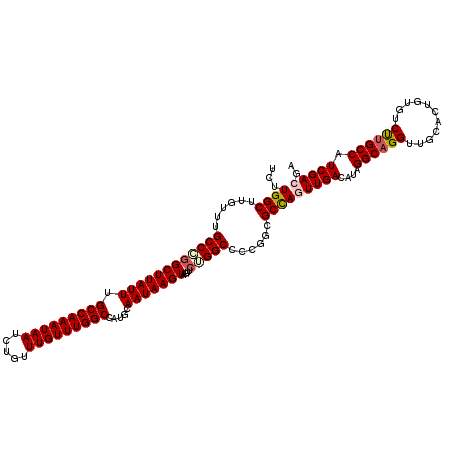
| Location | 16,556,528 – 16,556,648 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.82 |
| Mean single sequence MFE | -39.74 |
| Consensus MFE | -33.90 |
| Energy contribution | -34.62 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16556528 120 + 22407834 UCUUGGCUUGUUUGCCCGGCUUAUUUGCCAAAUAAUCUGUUUGUUUGGCCAUGCAAUAAGUAGUUCUGGCCCCGGCGCCAGUUGACAUAGGCAGGUUGCACUGUGCCUUGCCAUCGACGA ....(((......)))(((((((((.(((((((((.....))))))))).....))))))).((((((((......)))))..)))...((((((..((.....)))))))).....)). ( -37.50) >DroSec_CAF1 11054 120 + 1 UCUUGGCUUGUUUGCCCGGCUUAUUUGCCAAAUAAUCUGUUUGUUUGGCCAUGCAAUAAGUAGUUCGGGCCCCGGCGCCAGUUGACAUAGGCAGGUUGCACUGUGUCUUGCCAUCGACGA ...(((((((...((((((((((((.(((((((((.....))))))))).....)))))))....)))))..))).))))(((((....((((((.(((...))).)))))).))))).. ( -41.10) >DroSim_CAF1 11003 120 + 1 UCUUGGCUUGUUUGCCCGGCUUAUUUGCCAAAUAAUCUGUUUGUUUGGCCAUGCAAUAAGUAGUUCGGGCCCCGGCGCCAGUUGACAUAGGCAGGUUGCACUGUGUCUUGCCAUCGACGA ...(((((((...((((((((((((.(((((((((.....))))))))).....)))))))....)))))..))).))))(((((....((((((.(((...))).)))))).))))).. ( -41.10) >DroEre_CAF1 10896 117 + 1 UCUUGGCUUGUUUGCCCAGCUUAUUUGCCAAAUAAUCUGUUUGUUUGGCCAUGCAAUAAGUAGUUCUGGCCCCGUCGCCAGUUGACAUAGGCAGGUUGGACUGUGUCCCGCCAUCGA--- ...((((...((((((..(((((((.(((((((((.....))))))))).....))))))).((((((((......)))))..)))...))))))..((((...)))).))))....--- ( -38.10) >DroYak_CAF1 10863 117 + 1 UCUUGGCUUGUUUGCCAGGCUUAUUUGCCAAAUAAUCUGUUUGUUUGGCCAUGCAAUAAGUAGUUCUGGCCCCGGCGCUAGUUGACAUAGGCAGGUAGGACUGUGUCUUGCCAUCGA--- ..(((((......)))))((((((..(((((((((.....))))))))).......(((.((((.(((....))).)))).)))..)))))).((((((((...)))))))).....--- ( -40.90) >consensus UCUUGGCUUGUUUGCCCGGCUUAUUUGCCAAAUAAUCUGUUUGUUUGGCCAUGCAAUAAGUAGUUCUGGCCCCGGCGCCAGUUGACAUAGGCAGGUUGCACUGUGUCUUGCCAUCGACGA ...((((......((((((((((((.(((((((((.....))))))))).....)))))))....)))))......))))(((((....((((((...........)))))).))))).. (-33.90 = -34.62 + 0.72)

| Location | 16,556,528 – 16,556,648 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.82 |
| Mean single sequence MFE | -34.12 |
| Consensus MFE | -28.70 |
| Energy contribution | -29.22 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16556528 120 - 22407834 UCGUCGAUGGCAAGGCACAGUGCAACCUGCCUAUGUCAACUGGCGCCGGGGCCAGAACUACUUAUUGCAUGGCCAAACAAACAGAUUAUUUGGCAAAUAAGCCGGGCAAACAAGCCAAGA .......((((.(((((..........))))).((((..(((((......))))).....((((((.....((((((.((.....)).)))))).))))))...)))).....))))... ( -33.50) >DroSec_CAF1 11054 120 - 1 UCGUCGAUGGCAAGACACAGUGCAACCUGCCUAUGUCAACUGGCGCCGGGGCCCGAACUACUUAUUGCAUGGCCAAACAAACAGAUUAUUUGGCAAAUAAGCCGGGCAAACAAGCCAAGA .......((((..((((.((.((.....)))).))))..(((....))).(((((.....((((((.....((((((.((.....)).)))))).)))))).)))))......))))... ( -35.40) >DroSim_CAF1 11003 120 - 1 UCGUCGAUGGCAAGACACAGUGCAACCUGCCUAUGUCAACUGGCGCCGGGGCCCGAACUACUUAUUGCAUGGCCAAACAAACAGAUUAUUUGGCAAAUAAGCCGGGCAAACAAGCCAAGA .......((((..((((.((.((.....)))).))))..(((....))).(((((.....((((((.....((((((.((.....)).)))))).)))))).)))))......))))... ( -35.40) >DroEre_CAF1 10896 117 - 1 ---UCGAUGGCGGGACACAGUCCAACCUGCCUAUGUCAACUGGCGACGGGGCCAGAACUACUUAUUGCAUGGCCAAACAAACAGAUUAUUUGGCAAAUAAGCUGGGCAAACAAGCCAAGA ---....((((.((((...))))....((((((......(((((......))))).....((((((.....((((((.((.....)).)))))).)))))).)))))).....))))... ( -36.40) >DroYak_CAF1 10863 117 - 1 ---UCGAUGGCAAGACACAGUCCUACCUGCCUAUGUCAACUAGCGCCGGGGCCAGAACUACUUAUUGCAUGGCCAAACAAACAGAUUAUUUGGCAAAUAAGCCUGGCAAACAAGCCAAGA ---....((((..(((((((......)))....)))).......((((((..........((((((.....((((((.((.....)).)))))).))))))))))))......))))... ( -29.90) >consensus UCGUCGAUGGCAAGACACAGUGCAACCUGCCUAUGUCAACUGGCGCCGGGGCCAGAACUACUUAUUGCAUGGCCAAACAAACAGAUUAUUUGGCAAAUAAGCCGGGCAAACAAGCCAAGA .......((((..((((.((.((.....)))).))))..(((....))).(((((.....((((((.....((((((.((.....)).)))))).)))))).)))))......))))... (-28.70 = -29.22 + 0.52)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:15 2006