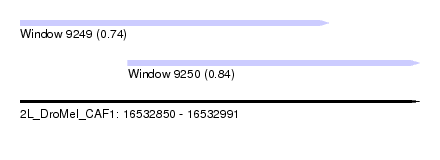
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,532,850 – 16,532,991 |
| Length | 141 |
| Max. P | 0.841196 |

| Location | 16,532,850 – 16,532,959 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 95.54 |
| Mean single sequence MFE | -17.27 |
| Consensus MFE | -16.26 |
| Energy contribution | -16.26 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
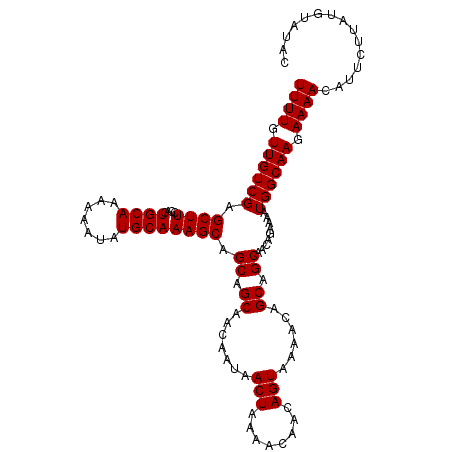
>2L_DroMel_CAF1 16532850 109 + 22407834 UUUUGUUGUCGAGCUUCCACUGCAAAAAAUAUGCAAAGCAGCAGCAACAAUAACUAAAACAACAGUAAAACAGCAGCAGCAGAAAAUGGCAAGAAAACAU---UACAUAUAC ((((.((((((...(((..((((........(((......)))((.......(((........)))......)).))))..)))..)))))).))))...---......... ( -17.52) >DroSec_CAF1 44451 112 + 1 UUUUGUUGUCGAGCUUCCACUGCAAAAAAUAUGCAAAGCAGCAGCAACAAUAACUAAAACAACAGUAAAACAGCAGCAACUGAAAAUGGCAAGAAAACAUUCUUAUGUAUAC .(((.((((((.((((....((((.......)))))))).((.((.......(((........)))......)).)).........)))))).)))((((....)))).... ( -16.52) >DroSim_CAF1 30960 112 + 1 UUUUGUUGUCGAGCUUCCACUGCAAAAAAUAUGCAAAGCAGCAGCAACAAUAACUAAAACAACAGUAAAACAGCAGCAACUGAAAAUGGCAAGAAAACAUUCUUAUGUAUAC .(((.((((((.((((....((((.......)))))))).((.((.......(((........)))......)).)).........)))))).)))((((....)))).... ( -16.52) >DroEre_CAF1 50386 112 + 1 UUUUGUUGUCGAGCUUCCACUGCAAAAAAUAUGCAAAGCAGCAGCAACAAUAACUAAAACAACAGUGAAACAGCAGCAACAGAAAAUGGCAAGAAAACAUUCUUAUGUAUAU (((((((((...(((..(((((((.......)))...((....))..................))))....))).)))))))))......(((((....)))))........ ( -17.90) >DroYak_CAF1 38369 112 + 1 UUUUGUUGUCGAGCUUCCACUGCAAAAAAUAUGCAAAGCAGCAGCAACAACAACUAAAACAACAGUGAAACAGCAGCAACAGAAAAUGGCAAGAAAACAUUCUUAUGUAUGU (((((((((...(((..(((((((.......)))...((....))..................))))....))).)))))))))......(((((....)))))........ ( -17.90) >consensus UUUUGUUGUCGAGCUUCCACUGCAAAAAAUAUGCAAAGCAGCAGCAACAAUAACUAAAACAACAGUAAAACAGCAGCAACAGAAAAUGGCAAGAAAACAUUCUUAUGUAUAC ((((.((((((.((((....((((.......)))))))).((.((.......(((........)))......)).)).........)))))).))))............... (-16.26 = -16.26 + -0.00)

| Location | 16,532,888 – 16,532,991 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 89.47 |
| Mean single sequence MFE | -19.31 |
| Consensus MFE | -15.47 |
| Energy contribution | -16.92 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
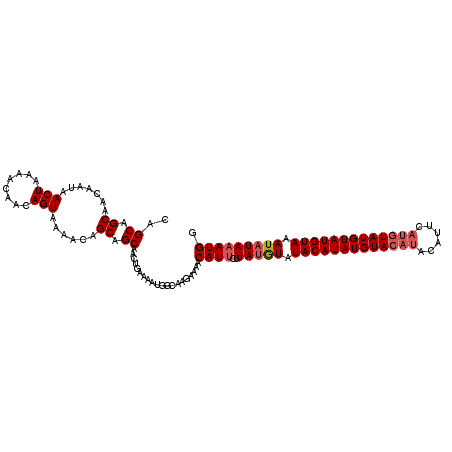
>2L_DroMel_CAF1 16532888 103 + 22407834 CAGCAGCAACAAUAACUAAAACAACAGUAAAACAGCAGCAGCAGAAAAUGGCAAGAAAACAU---UACAUAUACAUAUGUAUGUA--------UAUGUAUGUAAAUAUACAUGG ..((.((.......(((........)))......)).)).((........))......((((---.((((((((((....)))))--------)))))))))............ ( -16.52) >DroSec_CAF1 44489 114 + 1 CAGCAGCAACAAUAACUAAAACAACAGUAAAACAGCAGCAACUGAAAAUGGCAAGAAAACAUUCUUAUGUAUACAUAUGUACAUACAUUCAUGUAUGUAUGUAAAUAUAAAUGG .....((.......(((........)))....(((......)))......)).......((((..(((((.(((((((((((((......))))))))))))).))))))))). ( -20.70) >DroSim_CAF1 30998 114 + 1 CAGCAGCAACAAUAACUAAAACAACAGUAAAACAGCAGCAACUGAAAAUGGCAAGAAAACAUUCUUAUGUAUACAUAUGUACAUACAUUCAUGUAUGUAUGUAAAUAUAAAUGG .....((.......(((........)))....(((......)))......)).......((((..(((((.(((((((((((((......))))))))))))).))))))))). ( -20.70) >consensus CAGCAGCAACAAUAACUAAAACAACAGUAAAACAGCAGCAACUGAAAAUGGCAAGAAAACAUUCUUAUGUAUACAUAUGUACAUACAUUCAUGUAUGUAUGUAAAUAUAAAUGG ..((.((.......(((........)))......)).))....................((((..(((((.(((((((((((((......))))))))))))).))))))))). (-15.47 = -16.92 + 1.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:01 2006