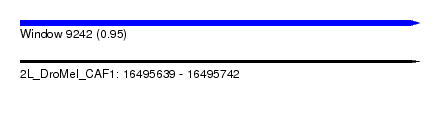
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,495,639 – 16,495,742 |
| Length | 103 |
| Max. P | 0.949937 |

| Location | 16,495,639 – 16,495,742 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.78 |
| Mean single sequence MFE | -27.05 |
| Consensus MFE | -14.74 |
| Energy contribution | -14.93 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.949937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16495639 103 + 22407834 ----ACACAU--------GGAAUUAAUGGGUCGGACAACGACCCAAAACUGUGG-AACAUAAAUUCAAUUGAGGCAAGUGUCAAUAGAACACGAG--C--UCAUCCAUUUGUAAGUGUUC ----(((.((--------(((.....(((((((.....))))))).........-..............((((.(..(((((....).))))..)--)--)))))))).)))........ ( -28.00) >DroPse_CAF1 8318 99 + 1 -------------------GCAAUAAUGGGUCGGACAACGACCCAAAACUGGGGCCUCAUAAAUUCAAUUGAGGCAAGUGUCAAUAGAACACGAGAAC--UCAUCCAUAUGCAAGCAUUC -------------------(((....(((((((.....)))))))....((((((((((..........))))))..(((((....).)))).....)--)))......)))........ ( -31.90) >DroEre_CAF1 20365 100 + 1 ------ACAU--------CGAA-UAAUGGGUCGGACAACGACCCAGAAGUGGGG-AACAUAAAUUCAAUUGAAGCAAGUGUCAAUAGAACACGAG--C--UCAUCCAUUUGCAAGUGUUC ------....--------.(((-((.(((((((.....))))))).((((((((-((......)))...(((.((..(((((....).))))..)--)--)))))))))).....))))) ( -28.30) >DroWil_CAF1 62091 104 + 1 UAGUAUCCAA---------GUAAUAAUGGUUCGAGCAAUGACCCAAAACUCGUA-AACAUAAAUUCAAUUGAAGCAAGUGUCAAUAGAACACGAA--C--UCUUCCAUUU-UACAUGUU- ..........---------((((.(((((...(((..((((........)))).-........(((.(((((........))))).)))......--)--))..))))))-))).....- ( -14.00) >DroAna_CAF1 14757 112 + 1 ------GCAUGCCGAGAGAGAAAUAAUAGGUCGGACAACGACCCAAAACUGGGGGGCCAUAAAUUCAAUUGAAGCAAGUGUCAAUAGAACACGAA--CUCUCAUCCAUAUGCAAGUUUUU ------(((((..(((((((........(((((.....)..((((....)))).)))).....(((.(((((........))))).)))......--))))).))..)))))........ ( -28.20) >DroPer_CAF1 8421 99 + 1 -------------------GCAAUAAUGGGUCGGACAACGACCCAAAACUGGGGCCUCAUAAAUUCAAUUGAGGCAAGUGUCAAUAGAACACGAGAAC--UCAUCCAUAUGCAAGCAUUC -------------------(((....(((((((.....)))))))....((((((((((..........))))))..(((((....).)))).....)--)))......)))........ ( -31.90) >consensus _______CAU_________GAAAUAAUGGGUCGGACAACGACCCAAAACUGGGG_AACAUAAAUUCAAUUGAAGCAAGUGUCAAUAGAACACGAG__C__UCAUCCAUAUGCAAGUGUUC ..........................(((((((.....)))))))..................(((.(((((.(....).))))).)))............................... (-14.74 = -14.93 + 0.19)

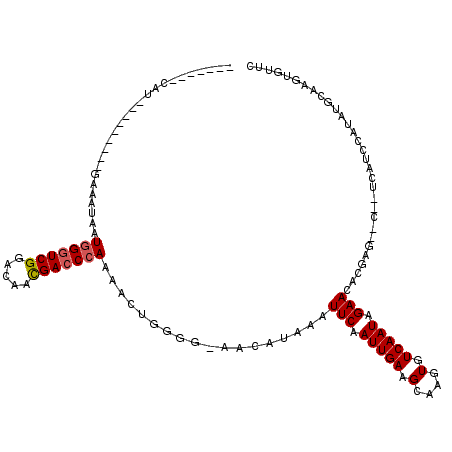
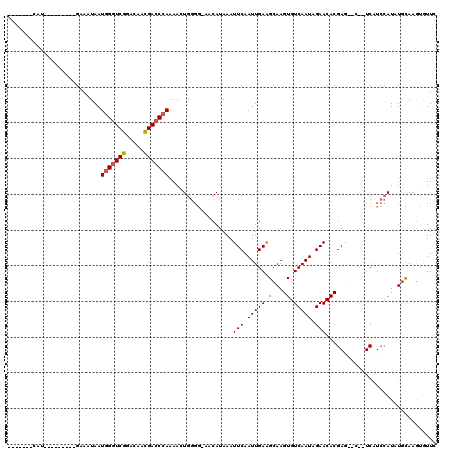
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:53 2006