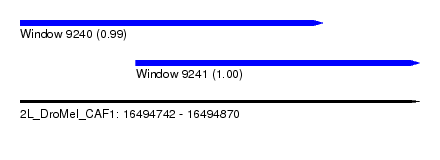
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,494,742 – 16,494,870 |
| Length | 128 |
| Max. P | 0.998221 |

| Location | 16,494,742 – 16,494,839 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -44.52 |
| Consensus MFE | -19.80 |
| Energy contribution | -22.11 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -5.29 |
| Structure conservation index | 0.44 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16494742 97 + 22407834 GCCAUUGUUGUUGGCGUAUCUGCUGCUCAUGUUGCUGUUGCAGUUGCAAUU-------------GCA-GUUGCUGUGGGCGA-GCAGAUUGCGUUUUAAAUUGCAGUGGCAU ((((((((.(((.(((((((((((((((((((.((((((((....)))...-------------)))-)).)).)))))).)-))))).)))))....))).)))))))).. ( -45.80) >DroEre_CAF1 19522 109 + 1 GCCAUUGUUGUUGGCGUAUCUGCUGCUCAUGUUGCAGUUGCAGUGGCAAUUUUUGUUGCAGUUG-UA-GUUUCUGUGGGCGG-GCAGAUUGCGUUUUAAAUUGCAGUGGCAU ((((((((.(((.(((((((((((((((((((((((..(((((..(......)..)))))..))-))-)....))))))).)-))))).)))))....))).)))))))).. ( -46.90) >DroWil_CAF1 59519 106 + 1 CCCCU-CCCGCUUGCCUCUUGGCUCUUGCUGCUGCUAUUGCAGCUUUAACUGCUGUG---GUUG-CAACUGGCUGUAGGCGUUGCAGAUUGCAU-UUAAAUUGCAGUGGCAU .....-...((..(((....)))....))...(((((((((((.(((((.(((....---.(((-((((.(.(....).))))))))...))).-)))))))))))))))). ( -36.70) >DroYak_CAF1 7604 109 + 1 GCCAUUGUUGUUGGCGUAUCUGCUGCUCAUGUUGCUGUUGCAGUUUCAAUUUUUGUUGCAGCUG-CA-GUUGCUGUGGGCGG-GAAGAUUGCGUUUUAAAUUGCAGUGGCAU ((((((((.(((.((((((((.((((((((...((..((((((((.((((....)))).)))))-))-)..)).))))))))-..))).)))))....))).)))))))).. ( -48.70) >consensus GCCAUUGUUGUUGGCGUAUCUGCUGCUCAUGUUGCUGUUGCAGUUGCAAUUUUUGUU___GUUG_CA_GUUGCUGUGGGCGG_GCAGAUUGCGUUUUAAAUUGCAGUGGCAU ((((((((.(((.((((((((((((((((((((((....)))))..............................)))))))..))))).)))))....))).)))))))).. (-19.80 = -22.11 + 2.31)


| Location | 16,494,779 – 16,494,870 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 80.93 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -21.04 |
| Energy contribution | -21.72 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.67 |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.998221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16494779 91 + 22407834 UUGCAGUUGCAAUU-------------GCA-GUUGCUGUGGGCGA-GCAGAUUGCGUUUUAAAUUGCAGUGGCAUUAAAUUUUGCAUGCAUUUUAUUGCAGUUGCA .((((..(((((..-------------(((-(((((((....).)-))).)))))....((((.((((...(((........))).)))).)))))))))..)))) ( -30.10) >DroEre_CAF1 19559 103 + 1 UUGCAGUGGCAAUUUUUGUUGCAGUUG-UA-GUUUCUGUGGGCGG-GCAGAUUGCGUUUUAAAUUGCAGUGGCAUUAAAUUUUGCAUGCACUUUAUUGCAGUUGCA ((((((..(((((....)))))..)))-))-)...(((....)))-(((.((((((...((((.((((...(((........))).)))).)))).))))))))). ( -30.30) >DroWil_CAF1 59555 101 + 1 UUGCAGCUUUAACUGCUGUG---GUUG-CAACUGGCUGUAGGCGUUGCAGAUUGCAU-UUAAAUUGCAGUGGCAUUAAAUUUUGUAUGCAUUUUAUUGCAGUUUGU ((((((((.....(((....---...)-))...)))))))).....((((((((((.-.((((.((((...(((........))).)))).)))).)))))))))) ( -32.50) >DroYak_CAF1 7641 103 + 1 UUGCAGUUUCAAUUUUUGUUGCAGCUG-CA-GUUGCUGUGGGCGG-GAAGAUUGCGUUUUAAAUUGCAGUGGCAUUAAAUUUUGCAUGCAUUUUAUUGCAGUUGCA .(((((((.((((....)))).)))))-))-((.((((..(((((-.....))))....((((.((((...(((........))).)))).)))))..)))).)). ( -33.00) >consensus UUGCAGUUGCAAUUUUUGUU___GUUG_CA_GUUGCUGUGGGCGG_GCAGAUUGCGUUUUAAAUUGCAGUGGCAUUAAAUUUUGCAUGCAUUUUAUUGCAGUUGCA ((((((((((.....................))))))))))........(((((((...((((.((((...(((........))).)))).)))).)))))))... (-21.04 = -21.72 + 0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:52 2006