
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
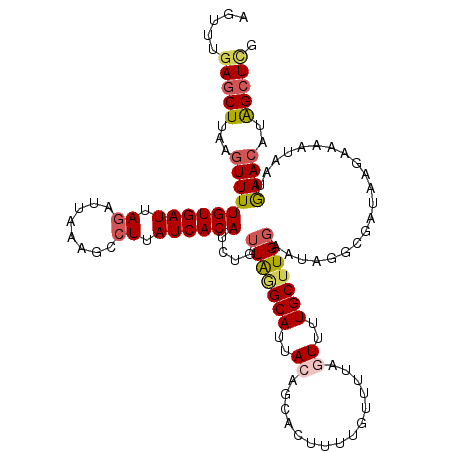
| Location | 16,490,237 – 16,490,349 |
| Length | 112 |
| Max. P | 0.733735 |

| Location | 16,490,237 – 16,490,349 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.91 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -14.00 |
| Energy contribution | -14.88 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.52 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16490237 112 + 22407834 AGUUUGAGCUUAAGUUUUGUGAUUAGAUUAAAGCCUUAUCACAUCUGUUAGGCAUUACAGCACUUUUGUUUUAGUUUUGCUUGGAAUAAG----AAGAAAA-AAGAAAAAGAGCUCG .....((((((......((((((.((........)).))))))(((.(((((((..((((((....))))...))..)))))))....))----)......-........)))))). ( -25.10) >DroSec_CAF1 2294 117 + 1 AGUUUGAGCUUAAGUUUUGUGAUAAGAUUAAAGCCUUAUCACAUCUGUUAGGCAUUACAGCACUUUUGUUUUAGUUUUGCUUGGAAUAGGCGAUAAGAAAAUAAUGAACAUAGCUCG .....(((((...(((((((((((((........))))))))).((((........)))).....(((((((....(((((((...)))))))....))))))).))))..))))). ( -29.10) >DroSim_CAF1 2277 117 + 1 AGUUUGAGCUUAAGUUUUGUGAUUAGAUUAAAGCCUUAUCACAUCUGUUAGGCAUUACAGCACUUUUGUUUUAGUUUUGCUUGGAAUAGGCGAUAAGAAAAUAAUGAACAUAGCUCG .....(((((...((((((((((.((........)).)))))).((((........)))).....(((((((....(((((((...)))))))....))))))).))))..))))). ( -25.10) >DroEre_CAF1 15082 112 + 1 AGUUUGAGCUUAUGUUUUGUGAUUAGAUGU---UAUGAUCACA--UGUUGAGCAUUACAGCUCUUAAAAAGCACUUUUGCUUGGAAUAGCUGAUAAGUAAAUAAUGAACAUAGCUAA ......((((.((((((((((((((.....---..))))))))--((((..((.((((((((.((...(((((....)))))..)).))))).))))).))))..)))))))))).. ( -29.50) >DroYak_CAF1 3024 115 + 1 AGCAUGAUCUUAGGUUUUGUGAUUAGAAUAAAGCCUUAUCACA--UGUUAUGCAUUACAGCACUUAAAAUAACUUUAUGCUUGUAAUGGCUAAUAAGAAAAUAAUGAACAUGGCUUG (((.((((...(((((((((.......))))))))).))))((--((((...((((((((((..((((.....))))))).))))))).((....)).........))))))))).. ( -26.60) >consensus AGUUUGAGCUUAAGUUUUGUGAUUAGAUUAAAGCCUUAUCACAUCUGUUAGGCAUUACAGCACUUUUGUUUUAGUUUUGCUUGGAAUAGGCGAUAAGAAAAUAAUGAACAUAGCUCG .....(((((...((((((((((.((........)).))))))....(((((((..((...............))..))))))).....................))))..))))). (-14.00 = -14.88 + 0.88)


| Location | 16,490,237 – 16,490,349 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.91 |
| Mean single sequence MFE | -20.88 |
| Consensus MFE | -10.88 |
| Energy contribution | -11.76 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16490237 112 - 22407834 CGAGCUCUUUUUCUU-UUUUCUU----CUUAUUCCAAGCAAAACUAAAACAAAAGUGCUGUAAUGCCUAACAGAUGUGAUAAGGCUUUAAUCUAAUCACAAAACUUAAGCUCAAACU .(((((..((((..(-(((.(((----........))).))))..))))...((((.((((........)))).((((((.(((......))).))))))..)))).)))))..... ( -18.40) >DroSec_CAF1 2294 117 - 1 CGAGCUAUGUUCAUUAUUUUCUUAUCGCCUAUUCCAAGCAAAACUAAAACAAAAGUGCUGUAAUGCCUAACAGAUGUGAUAAGGCUUUAAUCUUAUCACAAAACUUAAGCUCAAACU .(((((....................((.........)).............((((.((((........)))).((((((((((......))))))))))..)))).)))))..... ( -23.10) >DroSim_CAF1 2277 117 - 1 CGAGCUAUGUUCAUUAUUUUCUUAUCGCCUAUUCCAAGCAAAACUAAAACAAAAGUGCUGUAAUGCCUAACAGAUGUGAUAAGGCUUUAAUCUAAUCACAAAACUUAAGCUCAAACU .(((((....................((.........)).............((((.((((........)))).((((((.(((......))).))))))..)))).)))))..... ( -19.20) >DroEre_CAF1 15082 112 - 1 UUAGCUAUGUUCAUUAUUUACUUAUCAGCUAUUCCAAGCAAAAGUGCUUUUUAAGAGCUGUAAUGCUCAACA--UGUGAUCAUA---ACAUCUAAUCACAAAACAUAAGCUCAAACU ..(((((((((..........(((.(((((.....(((((....)))))......)))))))).........--((((((....---.......)))))).))))).))))...... ( -20.90) >DroYak_CAF1 3024 115 - 1 CAAGCCAUGUUCAUUAUUUUCUUAUUAGCCAUUACAAGCAUAAAGUUAUUUUAAGUGCUGUAAUGCAUAACA--UGUGAUAAGGCUUUAUUCUAAUCACAAAACCUAAGAUCAUGCU ..(((.(((.((...............((.((((((.((((.............))))))))))))......--((((((.(((......))).))))))........)).)))))) ( -22.82) >consensus CGAGCUAUGUUCAUUAUUUUCUUAUCACCUAUUCCAAGCAAAACUAAAACAAAAGUGCUGUAAUGCCUAACAGAUGUGAUAAGGCUUUAAUCUAAUCACAAAACUUAAGCUCAAACU .(((((.......................((((...((((...............))))..)))).........((((((.(((......))).)))))).......)))))..... (-10.88 = -11.76 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:50 2006