
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
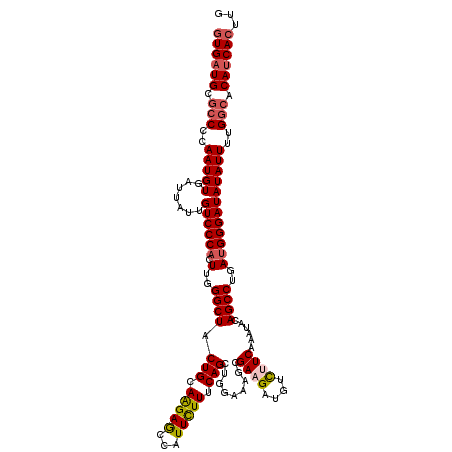
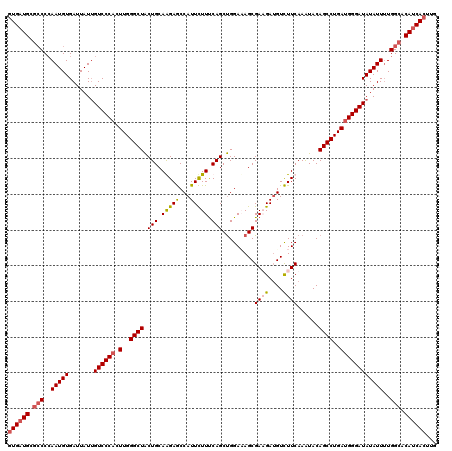
| Location | 16,485,531 – 16,485,651 |
| Length | 120 |
| Max. P | 0.977802 |

| Location | 16,485,531 – 16,485,651 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.53 |
| Mean single sequence MFE | -39.65 |
| Consensus MFE | -31.82 |
| Energy contribution | -32.76 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16485531 120 + 22407834 GUGAUGCGCCCCAAUGUGAUUAUUGUCCCCCUUGGGCUACUGCAAGAGCCAUUCUUUCAGCUGGAAAGCGAAGAUGUCUUCAAAUACAGCCUGAUGGGAUAUAUUAUGGCACAUCACUUG ((((((.(((.....(((((...((((((..(..((((..((.(((((...))))).))(((....)))((((....))))......))))..).)))))).)))))))).))))))... ( -41.80) >DroSec_CAF1 110520 120 + 1 GUGAUGCGGCCCAAUGUGAUUAUUGUCCCACUUGGGCUACUGCAGGAGCCAUUCUUUCAGCUGGACAGCGAAGAUGUCUUCAAAUACAGCCUGAUGGGAUAUAUUUUGGCACAUCACUUG ((((((..((((((((....)))))(((((.(..((((......((((.(((.((((..(((....)))))))))).))))......))))..))))))........))).))))))... ( -39.90) >DroSim_CAF1 107795 120 + 1 GUGAUGCGCCCCAAUGUGAUUAUUGUCCCACUUGGGCUACUGCAGGAGCCAUUCUUUCAGCUGGAAAGCGAUGAUGUCUUCAAGUACAGCCUGAUGGGAUAUAUUUUGGCACAUCACUUG ((((((.(((..(((((.......((((((.(..(((((((...((((.((((...((.(((....))))).)))).)))).)))..))))..))))))))))))..))).))))))... ( -44.61) >DroYak_CAF1 112574 120 + 1 AUGUUGCGCCCGAAUGUGAUUAUAGUCCCACUUGGGCUGCUGCAAGAACCAUUUUUCCAGCCGGAUAGCGAUGAAGUUUUCAAAUACAGCCUGAUGGGAUAUAUUUUGGGACAUCACUUG .((.((..((((((......((((.(((((.(..((((((((.((((.....)))).))))..((.(((......))).))......))))..)))))))))).)))))).)).)).... ( -32.30) >consensus GUGAUGCGCCCCAAUGUGAUUAUUGUCCCACUUGGGCUACUGCAAGAGCCAUUCUUUCAGCUGGAAAGCGAAGAUGUCUUCAAAUACAGCCUGAUGGGAUAUAUUUUGGCACAUCACUUG ((((((.(((..(((((.......((((((.(..((((.(((.(((((...))))).))).........((((....))))......))))..))))))))))))..))).))))))... (-31.82 = -32.76 + 0.94)

| Location | 16,485,531 – 16,485,651 |
|---|---|
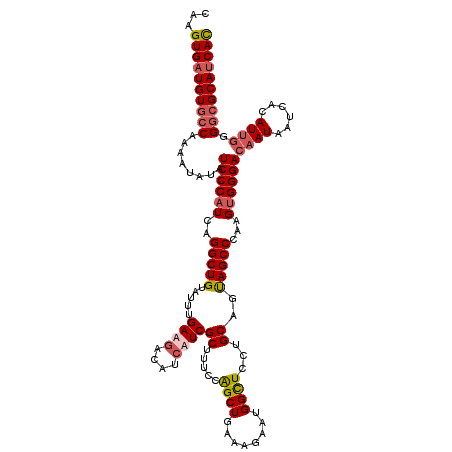
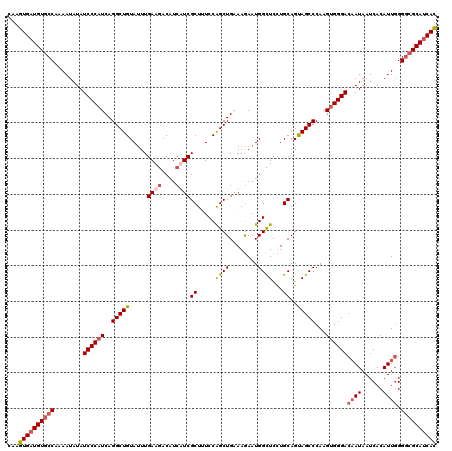
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.53 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -32.30 |
| Energy contribution | -33.80 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16485531 120 - 22407834 CAAGUGAUGUGCCAUAAUAUAUCCCAUCAGGCUGUAUUUGAAGACAUCUUCGCUUUCCAGCUGAAAGAAUGGCUCUUGCAGUAGCCCAAGGGGGACAAUAAUCACAUUGGGGCGCAUCAC ...((((((((((........((((....((((......((((....))))........((((.((((.....)))).))))))))....)))).((((......)))).)))))))))) ( -40.70) >DroSec_CAF1 110520 120 - 1 CAAGUGAUGUGCCAAAAUAUAUCCCAUCAGGCUGUAUUUGAAGACAUCUUCGCUGUCCAGCUGAAAGAAUGGCUCCUGCAGUAGCCCAAGUGGGACAAUAAUCACAUUGGGCCGCAUCAC ...((((((((((........((((((..((((......((((....))))(((((..((((........))))...)))))))))...))))))((((......)))))).)))))))) ( -41.50) >DroSim_CAF1 107795 120 - 1 CAAGUGAUGUGCCAAAAUAUAUCCCAUCAGGCUGUACUUGAAGACAUCAUCGCUUUCCAGCUGAAAGAAUGGCUCCUGCAGUAGCCCAAGUGGGACAAUAAUCACAUUGGGGCGCAUCAC ...((((((((((........((((((..(((((.............(((..(((((.....))))).)))((....))..)))))...))))))((((......)))).)))))))))) ( -39.10) >DroYak_CAF1 112574 120 - 1 CAAGUGAUGUCCCAAAAUAUAUCCCAUCAGGCUGUAUUUGAAAACUUCAUCGCUAUCCGGCUGGAAAAAUGGUUCUUGCAGCAGCCCAAGUGGGACUAUAAUCACAUUCGGGCGCAACAU ..(((((((.....(((((((.((.....)).)))))))........))))))).....((.((((......)))).)).((.((((.((((.((......)).)))).))))))..... ( -31.02) >consensus CAAGUGAUGUGCCAAAAUAUAUCCCAUCAGGCUGUAUUUGAAGACAUCAUCGCUUUCCAGCUGAAAGAAUGGCUCCUGCAGUAGCCCAAGUGGGACAAUAAUCACAUUGGGGCGCAUCAC ...((((((((((........((((((..(((((.....((((....))))((.....((((........))))...))..)))))...))))))((((......)))).)))))))))) (-32.30 = -33.80 + 1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:47 2006