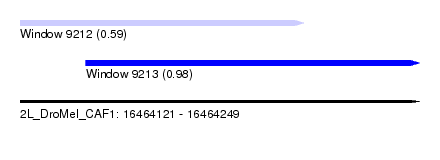
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,464,121 – 16,464,249 |
| Length | 128 |
| Max. P | 0.979852 |

| Location | 16,464,121 – 16,464,212 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 84.70 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -23.09 |
| Energy contribution | -23.40 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
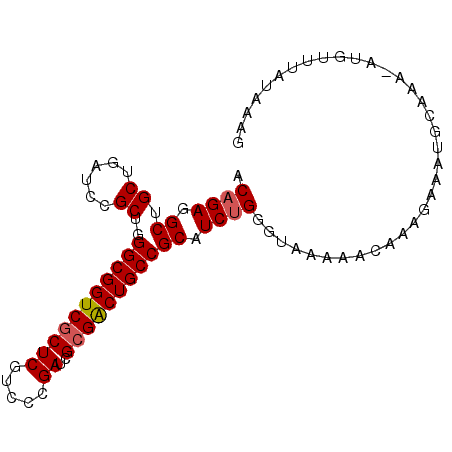
>2L_DroMel_CAF1 16464121 91 + 22407834 ACAGAGGCUGCUGAUCCGCUGGGCGGUCGCUCGUCCCGAUCGCGACUGCCGCAUCUGGGUGGAAACAAAGAAAUGGAAA-AUUUUUAUAGCA .((((.((.((......))..((((((((((((...)))..))))))))))).))))..((....)).((((((.....-))))))...... ( -29.80) >DroSec_CAF1 88833 91 + 1 ACAGAGGCUGCUGAUCCGCUGGGCGGUCGCUCGUCCCGAUCGCGACUGCCGCAUCUGGAUGGAAACAAAGAAAUGGAAA-AUGUUUAUAAAU .((((.((.((......))..((((((((((((...)))..))))))))))).))))..((....))............-............ ( -28.80) >DroEre_CAF1 86270 91 + 1 ACAGAGGCAGCUGAUCCGCUGGGCGGUCGCUCGUCCCGAUCGCGGCUGCCGCAUCUGGGUAAACAUAAGUAAGUGCAAA-AUGUUUAUCGAG .((((.(((((......))).((((((((((((...)))..))))))))))).))))(((((((((..((....))...-)))))))))... ( -35.10) >DroYak_CAF1 90990 91 + 1 ACUGAGGCUGCUGAUCCGCUGGGCGGUCUCUCAUCCCGAUCGCGACUGCCGCAUCUGGGAAAACAUA-GAAAAUGCAAAUAUUAUUGUAAAG ...(((((((((.(.....).)))))))))...(((((((.(((.....)))))).)))).......-.....(((((......)))))... ( -27.50) >consensus ACAGAGGCUGCUGAUCCGCUGGGCGGUCGCUCGUCCCGAUCGCGACUGCCGCAUCUGGGUAAAAACAAAGAAAUGCAAA_AUGUUUAUAAAG .((((.((.((......))..(((((((((((.....))..))))))))))).))))................................... (-23.09 = -23.40 + 0.31)

| Location | 16,464,142 – 16,464,249 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 68.21 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -16.42 |
| Energy contribution | -16.53 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
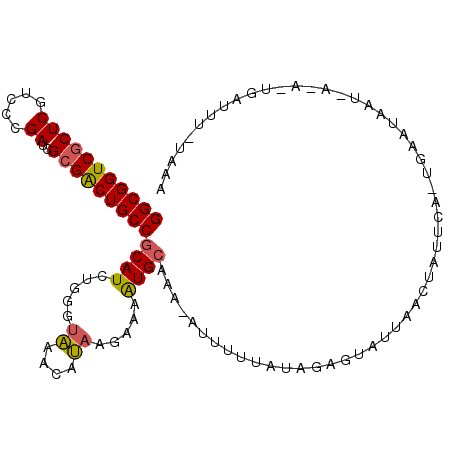
>2L_DroMel_CAF1 16464142 107 + 22407834 GGCGGUCGCUCGUCCCGAUCGCGACUGCCGCAUCUGGGUGGAAACAAAGAAAUGGAAA-AUUUUUAUAGCAUAUGAAUUAAUAAAUGAAUAAUUACACUGAUUU-UUCA ((((((((((((...)))..)))))))))...(((...((....)).)))..((((((-(((.........(((..(((....)))..)))........)))))-)))) ( -27.33) >DroEre_CAF1 86291 91 + 1 GGCGGUCGCUCGUCCCGAUCGCGGCUGCCGCAUCUGGGUAAACAUAAGUAAGUGCAAA-AUGUUUAUCGAGUAUUAUCUAUGCA-----------------UUUCUAAA ((((((((((((...)))..)))))))))((((((.(((((((((..((....))...-))))))))).))........)))).-----------------........ ( -29.80) >DroYak_CAF1 91011 106 + 1 GGCGGUCUCUCAUCCCGAUCGCGACUGCCGCAUCUGGGAAAACAUA-GAAAAUGCAAAUAUUAUUGUAAAGUGCUACCUAUUCAUUGAAUAAU-AUA-UGAUUUAUAAA (((((((.(((.....))..).)))))))(((((((........))-))...)))........(((((((........(((((...)))))..-...-...))))))). ( -19.29) >consensus GGCGGUCGCUCGUCCCGAUCGCGACUGCCGCAUCUGGGUAAACAUAAGAAAAUGCAAA_AUUUUUAUAGAGUAUUAACUAUUCA_UGAAUAAU_A_A_UGAUUU_UAAA (((((((((((.....))..)))))))))((((.....((....)).....))))...................................................... (-16.42 = -16.53 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:26 2006