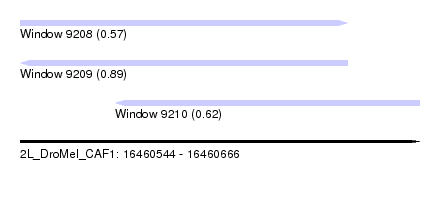
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,460,544 – 16,460,666 |
| Length | 122 |
| Max. P | 0.886727 |

| Location | 16,460,544 – 16,460,644 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -36.65 |
| Consensus MFE | -20.01 |
| Energy contribution | -21.78 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
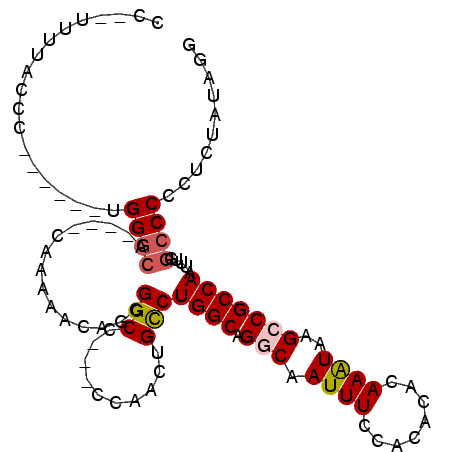
>2L_DroMel_CAF1 16460544 100 + 22407834 UUCGGCCGUC-UGGUCUAGGGACUAACUGCCCUAUAGAGGGGGCCAAAUUGGCGGCUUAUUUGUGUGUGGAAAUUGGCUGCCAGGCAGUU-----GG--GGGCCGCUG ..(((((.((-.((((....))))((((.((((....)))).(((....((((((((.((((.(....).)))).)))))))))))))))-----))--.)))))... ( -40.80) >DroSec_CAF1 85298 100 + 1 UUCGGCCGUC-UGGUCUAGGGACUAACUGCCCUAAAGAGGGGGCCAAAUUGGCGGCUUAUUUGUGUGUGGAAAUUGGCUGCCAGGCAGUU-----GG--GGGCCGCUG ..(((((.((-.((((....))))((((.((((....)))).(((....((((((((.((((.(....).)))).)))))))))))))))-----))--.)))))... ( -40.80) >DroEre_CAF1 82260 99 + 1 UUUGGCCGUC-UGGUCUAGGGACUAACUGCCCUAUGGAGGGGGCCAAAUUGGCGGCUUACUUGUGUGUGGAAAUUGCCUGCCAGGCAGUU-----GG---GGCCGCUG (((((((((.-(((((....)))))))..((((....)))))))))))..((((((((((......))...(((((((.....)))))))-----.)---))))))). ( -42.10) >DroYak_CAF1 87312 99 + 1 UUCGGCCGUC-UGGUCUAGGGAGUAUCUGCCCUAUGGAGGGGGCCAAAUUGGCGGCUUAUUUGUGUGUGGAAAUUGCCUGCCAGGCAGUU-----GG---GGCCGCUG ...((((.((-(..(((((((........))))).))))).)))).....((((((((..((((.((.((......))...)).))))..-----.)---))))))). ( -38.90) >DroAna_CAF1 78894 92 + 1 UUUGGCCUUUUUGGACUAUGG-------GCCU---AGUG-GGGGCAAAUUGGCGGCUUAUUUGUGUGUGGAAAUUGCCUGCCAGACUGUA-----GCCUUUGCCGUUG ...(((......((.((((((-------((((---....-.))))...((((((((..((((.(....).)))).))).))))).)))))-----)))...))).... ( -29.10) >DroPer_CAF1 58487 85 + 1 UCACGUUGUC-UGGUCUAGU-------------------GGGGGCAAAUUGGCGGCUUAUUUGUGUGUGGAAAUUGCCUGCCAAGCUGUUUUUUUGC---UGCCGCUG ..........-.....((((-------------------((.((((((..(((((((....((.(((.((......))))))))))))))..)))))---).)))))) ( -28.20) >consensus UUCGGCCGUC_UGGUCUAGGGACUAACUGCCCUAUAGAGGGGGCCAAAUUGGCGGCUUAUUUGUGUGUGGAAAUUGCCUGCCAGGCAGUU_____GG___GGCCGCUG ..(((((....((((((((((........)))).......))))))..(((((((...((((.(....).))))...)))))))................)))))... (-20.01 = -21.78 + 1.76)


| Location | 16,460,544 – 16,460,644 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -26.76 |
| Consensus MFE | -14.95 |
| Energy contribution | -17.07 |
| Covariance contribution | 2.13 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16460544 100 - 22407834 CAGCGGCCC--CC-----AACUGCCUGGCAGCCAAUUUCCACACACAAAUAAGCCGCCAAUUUGGCCCCCUCUAUAGGGCAGUUAGUCCCUAGACCA-GACGGCCGAA ...(((((.--..-----(((((((((((.......................)))(((.....)))..........)))))))).(((.........-)))))))).. ( -30.30) >DroSec_CAF1 85298 100 - 1 CAGCGGCCC--CC-----AACUGCCUGGCAGCCAAUUUCCACACACAAAUAAGCCGCCAAUUUGGCCCCCUCUUUAGGGCAGUUAGUCCCUAGACCA-GACGGCCGAA ...(((((.--..-----(((((((((((.......................)))(((.....)))..........)))))))).(((.........-)))))))).. ( -30.30) >DroEre_CAF1 82260 99 - 1 CAGCGGCC---CC-----AACUGCCUGGCAGGCAAUUUCCACACACAAGUAAGCCGCCAAUUUGGCCCCCUCCAUAGGGCAGUUAGUCCCUAGACCA-GACGGCCAAA ....((((---..-----(((((((((((.(((...................))))))....(((......)))..)))))))).(((.........-)))))))... ( -30.91) >DroYak_CAF1 87312 99 - 1 CAGCGGCC---CC-----AACUGCCUGGCAGGCAAUUUCCACACACAAAUAAGCCGCCAAUUUGGCCCCCUCCAUAGGGCAGAUACUCCCUAGACCA-GACGGCCGAA ..(((((.---..-----...((((.....))))((((........))))..)))))...(((((((..((...(((((.((...)))))))....)-)..))))))) ( -28.70) >DroAna_CAF1 78894 92 - 1 CAACGGCAAAGGC-----UACAGUCUGGCAGGCAAUUUCCACACACAAAUAAGCCGCCAAUUUGCCCC-CACU---AGGC-------CCAUAGUCCAAAAAGGCCAAA ....(((((((((-----....)))((((.(((...................))))))).))))))..-....---.(((-------(.............))))... ( -24.13) >DroPer_CAF1 58487 85 - 1 CAGCGGCA---GCAAAAAAACAGCUUGGCAGGCAAUUUCCACACACAAAUAAGCCGCCAAUUUGCCCCC-------------------ACUAGACCA-GACAACGUGA .((.((..---(((((........(((((.(((...................)))))))))))))..))-------------------.))......-.......... ( -16.21) >consensus CAGCGGCC___CC_____AACUGCCUGGCAGGCAAUUUCCACACACAAAUAAGCCGCCAAUUUGGCCCCCUCUAUAGGGCAGUUAGUCCCUAGACCA_GACGGCCGAA ..(((((.............((((...))))...((((........))))..)))))...(((((((.......(((((........))))).........))))))) (-14.95 = -17.07 + 2.13)
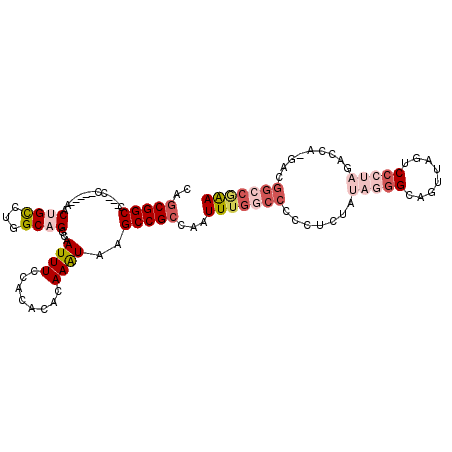

| Location | 16,460,573 – 16,460,666 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.81 |
| Mean single sequence MFE | -24.35 |
| Consensus MFE | -12.92 |
| Energy contribution | -13.40 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16460573 93 - 22407834 CC--UUUUACCC------UGGGCA----CAAAAACAGCGGCCC--CCAACUGCCUGGCAGCCAAUUUCCACACACAAAUAAGCCGCCAAUUUGGCCCCCUCUAUAGG ((--(.......------.((((.----((((......(((..--......)))((((.((..((((........))))..)).)))).)))))))).......))) ( -22.46) >DroSec_CAF1 85327 92 - 1 CC--UUUUACCC------UGGGCA----CAA-AACAGCGGCCC--CCAACUGCCUGGCAGCCAAUUUCCACACACAAAUAAGCCGCCAAUUUGGCCCCCUCUUUAGG ((--(.......------.((((.----(((-(.....(((..--......)))((((.((..((((........))))..)).)))).)))))))).......))) ( -22.46) >DroEre_CAF1 82289 94 - 1 CCAGUUUACCCC------UGGGCA----CAAGAACAGCGGCC---CCAACUGCCUGGCAGGCAAUUUCCACACACAAGUAAGCCGCCAAUUUGGCCCCCUCCAUAGG ............------.((((.----((((....(((((.---.....((((.....))))......((......))..)))))...))))))))(((....))) ( -23.60) >DroYak_CAF1 87341 92 - 1 CC--UUAUACCC------UGGCCA----GAAGUACAGCGGCC---CCAACUGCCUGGCAGGCAAUUUCCACACACAAAUAAGCCGCCAAUUUGGCCCCCUCCAUAGG ((--(.......------.(((((----((......((((..---....)))).((((.(((...................))))))).)))))))........))) ( -24.80) >DroAna_CAF1 78917 101 - 1 CC--UUCCACCUUUCAACGGGGCAGCCGCAGAAGCAACGGCAAAGGCUACAGUCUGGCAGGCAAUUUCCACACACAAAUAAGCCGCCAAUUUGCCCC-CACU---AG ..--..............((((((((((.........))))..((((....))))(((.(((...................))))))....))))))-....---.. ( -28.41) >consensus CC__UUUUACCC______UGGGCA____CAAAAACAGCGGCC___CCAACUGCCUGGCAGGCAAUUUCCACACACAAAUAAGCCGCCAAUUUGGCCCCCUCUAUAGG ...................((((...............(((..........)))((((.(((.((((........))))..))))))).....)))).......... (-12.92 = -13.40 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:24 2006