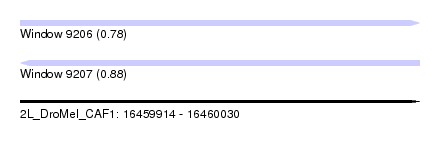
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,459,914 – 16,460,030 |
| Length | 116 |
| Max. P | 0.878950 |

| Location | 16,459,914 – 16,460,030 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -23.72 |
| Consensus MFE | -15.90 |
| Energy contribution | -15.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
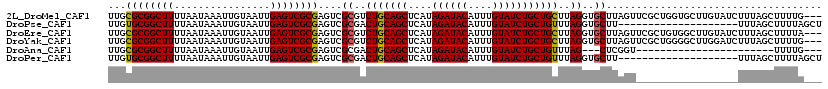
>2L_DroMel_CAF1 16459914 116 + 22407834 ---CAAAAGCUAAAGAUACAAGCACCAGCGAACUAAGCACCUAAGCAGCAGAUACAAAUGUAUCUAUGAGCUGCAGACGCGACUCGCGACUCAAUUACAAUUUAUUAAAAGCCGCGCAA ---.....(((.........)))....(((......((......(((((((((((....))))))....)))))...((((...))))......................))..))).. ( -24.90) >DroPse_CAF1 59688 99 + 1 AGCUAAAAGCUAAA--------------------AAGCACCUAAACAGCAGAUACAAAUGUAUCUAUGAGCUGCAGUCGCGACUCGCGACUCAAUUACAAUUUAUUAAAAGCCGCACAA (((.....)))...--------------------..((.......((((((((((....))))))....)))).(((((((...)))))))......................)).... ( -23.30) >DroEre_CAF1 81648 116 + 1 ---UAAAAGCUAAAGAUACAAGCCACAGCGAACUAAGCACCUAAGCAGCAGAUACAAAUGUAUCUAUGAGCUGCAGACGCGACUCGCGACUCAAUUACAAUUUAUUAAAAGCCGCGCAA ---.....(((.........)))....(((......((......(((((((((((....))))))....)))))...((((...))))......................))..))).. ( -24.00) >DroYak_CAF1 86675 116 + 1 ---CAAAAGCUAAAGAUCCAAGCCCCAGCGAACUAAGCACCUAAGCAGCAGAUACAAAUGUAUCUAUGAGCUGCAGACGCGACUCGCGACUCAAUUACAAUUUAUUAAAAGCCGCGCAA ---.....(((.........)))....(((......((......(((((((((((....))))))....)))))...((((...))))......................))..))).. ( -24.00) >DroAna_CAF1 78478 90 + 1 ---CAAAA-----------------------ACCGAG---CUAAACAGCAGAUACAAAUGUAUCUAUGAGCUGCAGUCGCGACUCGCGACUCAAUUACAAUUUAUUAAAAGCCGCGCAA ---.....-----------------------..((.(---((...((((((((((....))))))....)))).(((((((...)))))))..................)))))..... ( -22.80) >DroPer_CAF1 57929 99 + 1 AGCUAAAAGCUAAA--------------------AAGCACCUAAACAGCAGAUACAAAUGUAUCUAUGAGCUGCAGUCGCGACUCGCGACUCAAUUACAAUUUAUUAAAAGCCGCACAA (((.....)))...--------------------..((.......((((((((((....))))))....)))).(((((((...)))))))......................)).... ( -23.30) >consensus ___CAAAAGCUAAA_________________ACUAAGCACCUAAACAGCAGAUACAAAUGUAUCUAUGAGCUGCAGACGCGACUCGCGACUCAAUUACAAUUUAUUAAAAGCCGCGCAA ...............................................((((((((....)))))).((((.(((((......)).))).))))....................)).... (-15.90 = -15.90 + 0.00)

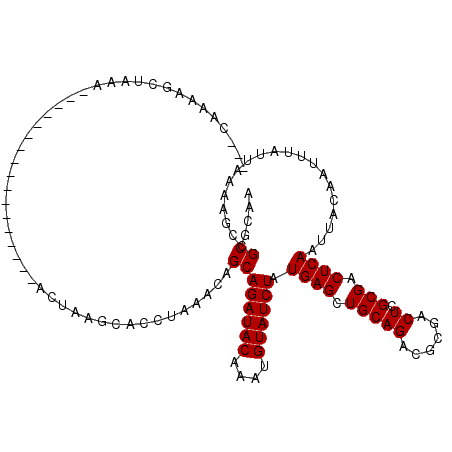
| Location | 16,459,914 – 16,460,030 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -25.56 |
| Energy contribution | -25.26 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.878950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16459914 116 - 22407834 UUGCGCGGCUUUUAAUAAAUUGUAAUUGAGUCGCGAGUCGCGUCUGCAGCUCAUAGAUACAUUUGUAUCUGCUGCUUAGGUGCUUAGUUCGCUGGUGCUUGUAUCUUUAGCUUUUG--- ...((((((((................))))))))....((....(((((....((((((....))))))))))).((((..((.((....))))..))))........)).....--- ( -33.19) >DroPse_CAF1 59688 99 - 1 UUGUGCGGCUUUUAAUAAAUUGUAAUUGAGUCGCGAGUCGCGACUGCAGCUCAUAGAUACAUUUGUAUCUGCUGUUUAGGUGCUU--------------------UUUAGCUUUUAGCU ......((((.....((((..(((....(((((((...)))))))(((((....((((((....))))))))))).....)))..--------------------))))......)))) ( -28.00) >DroEre_CAF1 81648 116 - 1 UUGCGCGGCUUUUAAUAAAUUGUAAUUGAGUCGCGAGUCGCGUCUGCAGCUCAUAGAUACAUUUGUAUCUGCUGCUUAGGUGCUUAGUUCGCUGUGGCUUGUAUCUUUAGCUUUUA--- ..(((((((((................)))))))(((((((....(((((....((((((....)))))))))))...((((.......))))))))))).........)).....--- ( -34.29) >DroYak_CAF1 86675 116 - 1 UUGCGCGGCUUUUAAUAAAUUGUAAUUGAGUCGCGAGUCGCGUCUGCAGCUCAUAGAUACAUUUGUAUCUGCUGCUUAGGUGCUUAGUUCGCUGGGGCUUGGAUCUUUAGCUUUUG--- ..(((((((((................)))))))(((((((..(((((((....((((((....)))))))))))..))..))((((....))))))))).........)).....--- ( -34.79) >DroAna_CAF1 78478 90 - 1 UUGCGCGGCUUUUAAUAAAUUGUAAUUGAGUCGCGAGUCGCGACUGCAGCUCAUAGAUACAUUUGUAUCUGCUGUUUAG---CUCGGU-----------------------UUUUG--- ...((((((((................))))))))......(((((.(((((((((((((....))))))).))...))---))))))-----------------------)....--- ( -26.09) >DroPer_CAF1 57929 99 - 1 UUGUGCGGCUUUUAAUAAAUUGUAAUUGAGUCGCGAGUCGCGACUGCAGCUCAUAGAUACAUUUGUAUCUGCUGUUUAGGUGCUU--------------------UUUAGCUUUUAGCU ......((((.....((((..(((....(((((((...)))))))(((((....((((((....))))))))))).....)))..--------------------))))......)))) ( -28.00) >consensus UUGCGCGGCUUUUAAUAAAUUGUAAUUGAGUCGCGAGUCGCGACUGCAGCUCAUAGAUACAUUUGUAUCUGCUGCUUAGGUGCUUAGU_________________UUUAGCUUUUA___ ...((((((((................))))))))....((..(((((((....((((((....)))))))))))..))..)).................................... (-25.56 = -25.26 + -0.30)
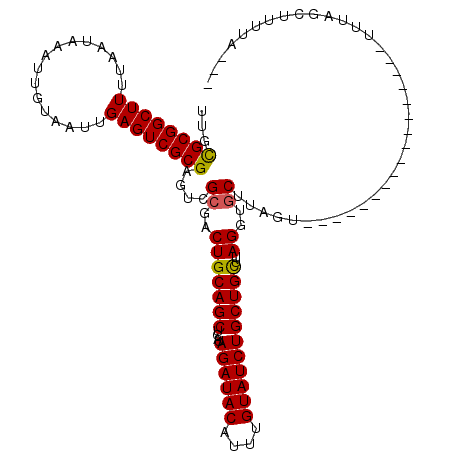
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:21 2006