| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,456,119 – 16,456,219 |
| Length | 100 |
| Max. P | 0.968859 |

| Location | 16,456,119 – 16,456,219 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.19 |
| Mean single sequence MFE | -13.88 |
| Consensus MFE | -12.33 |
| Energy contribution | -12.50 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16456119 100 + 22407834 -----------------AAAAACAUGUUAACCAGAAAAUUAAUUAUAAAAUUUUCA--GAAACCUUAACAAUGUCUACAAUAUGGCGUUGAUUUUACUCAAACAAA-AACUAACUAACUG -----------------.......((((.....(((((((........))))))).--..........(((((((........)))))))..........))))..-............. ( -13.40) >DroSim_CAF1 81077 100 + 1 -----------------AAAAACAUGUUAACCAGAAAAUUAAUUAUAAAAUUUUCA--GAAACCUUAACAAUGUCUACAAUAUGGCGUUGAUUUUACUCAAACAAA-AACUAACUAACUG -----------------.......((((.....(((((((........))))))).--..........(((((((........)))))))..........))))..-............. ( -13.40) >DroEre_CAF1 77953 100 + 1 -----------------AAAAACAUGUUAACCAGAAAAUUAAUUAUAAAAUUUUCA--GAAACCUUAACAAUGUCUACAAUACGGCGUUGAUUUUACUCAAACAAA-AACUAACUAACUG -----------------.......((((.....(((((((........))))))).--..........(((((((........)))))))..........))))..-............. ( -12.80) >DroWil_CAF1 105982 120 + 1 AUGUUCGUGUGUAUGGGAAAAACAUGUUAACCAGAAAAUUAAUUAUAAAAUUUUCACAAAAACCUUAACAAUGUCUACAAUAUGGCGUUGAUUUUACUCAAACAAAAAACUAACUAACUG ..(((.((.(((.((((((((............(((((((........))))))).............(((((((........))))))).)))).)))).)))....)).)))...... ( -20.20) >DroYak_CAF1 81990 100 + 1 -----------------AAAAACAUGUUAACCAGAAAAUUAAUUAUAAAAUUUUCA--GAAUCCUUAACAAUGUCUACAAUACGGCGUUGAUUUUACUCAAACAAA-AACUAACUAACUG -----------------.......((((.....(((((((........))))))).--..........(((((((........)))))))..........))))..-............. ( -12.80) >DroMoj_CAF1 51422 100 + 1 -----------------AAAAACAUGUUAACCAGAAAAUUAAUUAUAAAAUUUUCA--AAAACCUUAACAAUGUCUACAAUAUGGCGUUUAUUUUACUCAAACAAG-CACCAACUAACUG -----------------....(((((((((...(((((((........))))))).--......))))).))))........(((.((((.(((.....))).)))-).)))........ ( -10.70) >consensus _________________AAAAACAUGUUAACCAGAAAAUUAAUUAUAAAAUUUUCA__GAAACCUUAACAAUGUCUACAAUAUGGCGUUGAUUUUACUCAAACAAA_AACUAACUAACUG ........................((((.....(((((((........))))))).............(((((((........)))))))..........))))................ (-12.33 = -12.50 + 0.17)

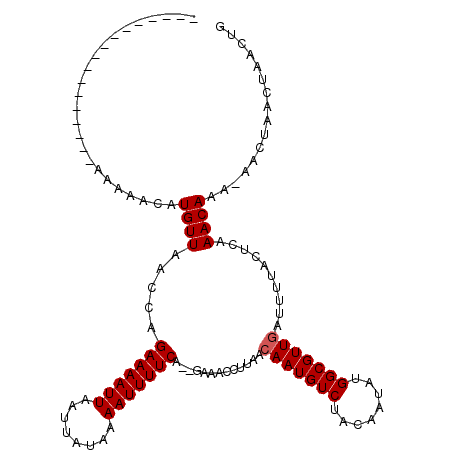

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:18 2006