
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,453,603 – 16,453,740 |
| Length | 137 |
| Max. P | 0.992975 |

| Location | 16,453,603 – 16,453,707 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.69 |
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -9.23 |
| Energy contribution | -9.87 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.35 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16453603 104 + 22407834 CUUUUGGGGAGAAUUGGUUUAUUUGAUCUACAAAUGGCUUAAGCUCAAAAGAGAGCUCUUUACUUAAUGUUGACUAUCAAA--------UGCGUUAAAGAGCUCU--------AUAUAUU ((((((((.....((((.(((((((.....))))))).)))).))))))))(((((((((((.......(((.....))).--------.....)))))))))))--------....... ( -26.92) >DroSec_CAF1 78335 103 + 1 CUU-UGGGGAGAAUUGGUUUAUUUGAUCUACAAAUGGCCUAAGCUCGAAAGAGAGCUCUUUACUUAAUUUUGAUUAUCAAA--------UGCGUUAAAGAGCUCU--------AUAAAUU (((-(.(((....((((..((((((.....))))))..)))).))).))))(((((((((((......((((.....))))--------.....)))))))))))--------....... ( -29.70) >DroSim_CAF1 78596 103 + 1 CUU-UGGGGAGAAUUGGUUUAUUUGAUCUACAAAUGGCCUAAGCUCGAAAGAGAGCUCUUUACUUAAUUUUGAUUAUCAAA--------UGCCUUGAAGAGCUCU--------AUAAAUU (((-(.(((....((((..((((((.....))))))..)))).))).))))(((((((((((......((((.....))))--------.....)))))))))))--------....... ( -29.80) >DroEre_CAF1 75196 98 + 1 ------------AUUGGUUUU-UUUAUUUACAAGUGCUAUGAGCUCGAAGGAGAGCUAUUUAGUUAAUUUUCAUUAUCAUA--------UGUGUGAAA-AGCUCUCGAGCAAAAUGUUUU ------------.........-.......(((..((((..(((((.(((((..((((....))))..)))))....((((.--------...))))..-)))))...))))...)))... ( -19.40) >DroYak_CAF1 79324 119 + 1 CUU-UGGGGAGAGUUGGUUUCCUUGAUUUACAAAUGGAAUGAGCUCGAAAAAAAGCUCUUUCUUUUAUUUUGAUUAUCAUAUGUACAUAUGUGUGAAAUAACUCUCGAGCUAAAUAUAUU .((-(((.((((((((.......(((((.......((((.(((((........))))).))))........)))))((((((((....))))))))..))))))))...)))))...... ( -26.86) >consensus CUU_UGGGGAGAAUUGGUUUAUUUGAUCUACAAAUGGCAUAAGCUCGAAAGAGAGCUCUUUACUUAAUUUUGAUUAUCAAA________UGCGUUAAAGAGCUCU________AUAUAUU .............((((((((..(.((......)).)..))))).)))...(((((((((((................................)))))))))))............... ( -9.23 = -9.87 + 0.64)

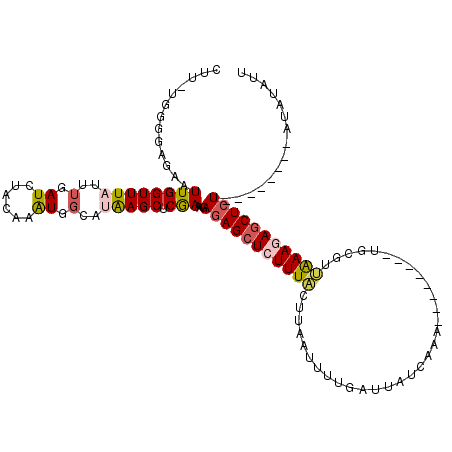
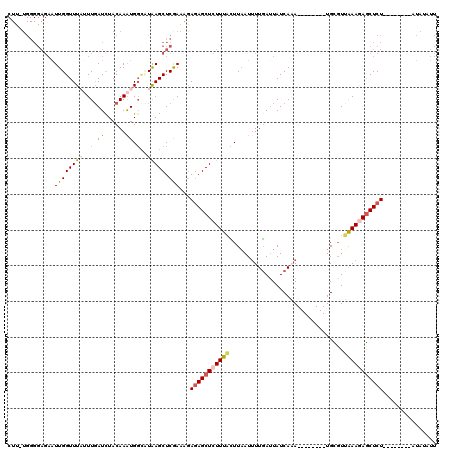
| Location | 16,453,603 – 16,453,707 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.69 |
| Mean single sequence MFE | -20.06 |
| Consensus MFE | -8.67 |
| Energy contribution | -8.91 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16453603 104 - 22407834 AAUAUAU--------AGAGCUCUUUAACGCA--------UUUGAUAGUCAACAUUAAGUAAAGAGCUCUCUUUUGAGCUUAAGCCAUUUGUAGAUCAAAUAAACCAAUUCUCCCCAAAAG .......--------(((((((((((.....--------.(((.....))).......)))))))))))((((((.((....)).(((((.....)))))..............)))))) ( -21.12) >DroSec_CAF1 78335 103 - 1 AAUUUAU--------AGAGCUCUUUAACGCA--------UUUGAUAAUCAAAAUUAAGUAAAGAGCUCUCUUUCGAGCUUAGGCCAUUUGUAGAUCAAAUAAACCAAUUCUCCCCA-AAG .......--------(((((((((((.....--------((((.....))))......))))))))))).....(((..(.((..(((((.....)))))...)).)..)))....-... ( -21.40) >DroSim_CAF1 78596 103 - 1 AAUUUAU--------AGAGCUCUUCAAGGCA--------UUUGAUAAUCAAAAUUAAGUAAAGAGCUCUCUUUCGAGCUUAGGCCAUUUGUAGAUCAAAUAAACCAAUUCUCCCCA-AAG .......--------(((((((((.......--------((((.....))))........))))))))).....(((..(.((..(((((.....)))))...)).)..)))....-... ( -17.76) >DroEre_CAF1 75196 98 - 1 AAAACAUUUUGCUCGAGAGCU-UUUCACACA--------UAUGAUAAUGAAAAUUAACUAAAUAGCUCUCCUUCGAGCUCAUAGCACUUGUAAAUAAA-AAAACCAAU------------ ..........(((((((((((-..(((....--------..)))((((....)))).......)))))))....))))....................-.........------------ ( -15.90) >DroYak_CAF1 79324 119 - 1 AAUAUAUUUAGCUCGAGAGUUAUUUCACACAUAUGUACAUAUGAUAAUCAAAAUAAAAGAAAGAGCUUUUUUUCGAGCUCAUUCCAUUUGUAAAUCAAGGAAACCAACUCUCCCCA-AAG ..............(((((((.((((...(((((....))))).........(((((.(((.((((((......)))))).)))..)))))........))))..)))))))....-... ( -24.10) >consensus AAUAUAU________AGAGCUCUUUAACGCA________UUUGAUAAUCAAAAUUAAGUAAAGAGCUCUCUUUCGAGCUUAGGCCAUUUGUAGAUCAAAUAAACCAAUUCUCCCCA_AAG ...............(((((((((((................................)))))))))))................................................... ( -8.67 = -8.91 + 0.24)

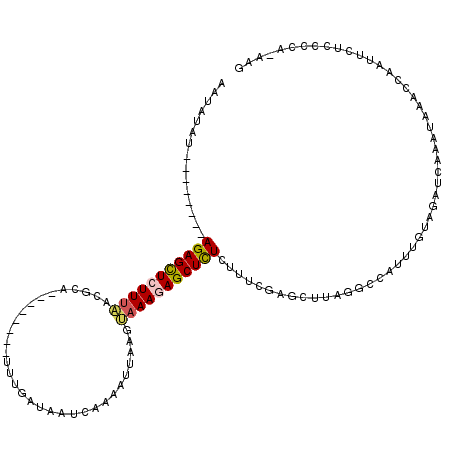
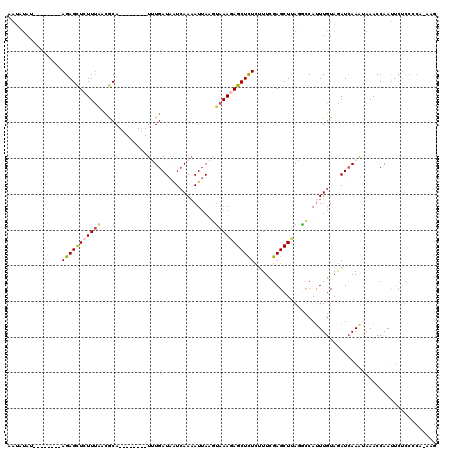
| Location | 16,453,643 – 16,453,740 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.86 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -9.77 |
| Energy contribution | -10.13 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16453643 97 + 22407834 AAGCUCAAAAGAGAGCUCUUUACUUAAUGUUGACUAUCAAA--------UGCGUUAAAGAGCUCU--------AUAUAUUAGGCCUUAAGUAACUGCUAAGCCAAUUAUUGCA------- ..((.......(((((((((((.......(((.....))).--------.....)))))))))))--------........(((.(((.((....)))))))).......)).------- ( -22.92) >DroSec_CAF1 78374 97 + 1 AAGCUCGAAAGAGAGCUCUUUACUUAAUUUUGAUUAUCAAA--------UGCGUUAAAGAGCUCU--------AUAAAUUAGGUCUUAAGUAACUGCUAAGCGAAUUACUGCA------- .(((((....))((((((((((......((((.....))))--------.....)))))))))).--------......................)))..(((......))).------- ( -22.40) >DroSim_CAF1 78635 97 + 1 AAGCUCGAAAGAGAGCUCUUUACUUAAUUUUGAUUAUCAAA--------UGCCUUGAAGAGCUCU--------AUAAAUUAGGCCUUAAGUAAUUGCUAAGCGAAUUACUGCA------- ..(((..(...(((((((((((......((((.....))))--------.....)))))))))))--------.....)..)))....(((((((((...)).)))))))...------- ( -24.80) >DroEre_CAF1 75223 110 + 1 GAGCUCGAAGGAGAGCUAUUUAGUUAAUUUUCAUUAUCAUA--------UGUGUGAAA-AGCUCUCGAGCAAAAUGUUUUAGGC-UUAAGUGAGUGCUUAGCAAAUUGCUGCUCAUUAUU ((((......(((((((...........((((((.......--------...))))))-))))))).(((((..(((..(((((-..........))))))))..)))))))))...... ( -28.40) >DroYak_CAF1 79363 113 + 1 GAGCUCGAAAAAAAGCUCUUUCUUUUAUUUUGAUUAUCAUAUGUACAUAUGUGUGAAAUAACUCUCGAGCUAAAUAUAUUAGGCCUUAAGUAAGACCUAAUCAAAUAACUGCA------- .(((((((..(((((......)))))..........((((((((....))))))))........)))))))......((((((.(((....))).))))))............------- ( -22.50) >consensus AAGCUCGAAAGAGAGCUCUUUACUUAAUUUUGAUUAUCAAA________UGCGUUAAAGAGCUCU________AUAUAUUAGGCCUUAAGUAACUGCUAAGCAAAUUACUGCA_______ ..((.......(((((((((((................................))))))))))).......................(((((............)))))))........ ( -9.77 = -10.13 + 0.36)



| Location | 16,453,643 – 16,453,740 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.86 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -9.41 |
| Energy contribution | -10.97 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16453643 97 - 22407834 -------UGCAAUAAUUGGCUUAGCAGUUACUUAAGGCCUAAUAUAU--------AGAGCUCUUUAACGCA--------UUUGAUAGUCAACAUUAAGUAAAGAGCUCUCUUUUGAGCUU -------.((.......(((((((......))..)))))........--------(((((((((((.....--------.(((.....))).......))))))))))).......)).. ( -21.92) >DroSec_CAF1 78374 97 - 1 -------UGCAGUAAUUCGCUUAGCAGUUACUUAAGACCUAAUUUAU--------AGAGCUCUUUAACGCA--------UUUGAUAAUCAAAAUUAAGUAAAGAGCUCUCUUUCGAGCUU -------.(((((((((.((...)))))))))...............--------(((((((((((.....--------((((.....))))......))))))))))).......)).. ( -22.80) >DroSim_CAF1 78635 97 - 1 -------UGCAGUAAUUCGCUUAGCAAUUACUUAAGGCCUAAUUUAU--------AGAGCUCUUCAAGGCA--------UUUGAUAAUCAAAAUUAAGUAAAGAGCUCUCUUUCGAGCUU -------((((((.....)))..))).((((((((.((((.......--------((....))...)))).--------((((.....)))).)))))))).((((((......)))))) ( -20.00) >DroEre_CAF1 75223 110 - 1 AAUAAUGAGCAGCAAUUUGCUAAGCACUCACUUAA-GCCUAAAACAUUUUGCUCGAGAGCU-UUUCACACA--------UAUGAUAAUGAAAAUUAACUAAAUAGCUCUCCUUCGAGCUC ......(((((((((..((.((.((..........-)).))...))..))))).(((((((-..(((....--------..)))((((....)))).......)))))))......)))) ( -21.80) >DroYak_CAF1 79363 113 - 1 -------UGCAGUUAUUUGAUUAGGUCUUACUUAAGGCCUAAUAUAUUUAGCUCGAGAGUUAUUUCACACAUAUGUACAUAUGAUAAUCAAAAUAAAAGAAAGAGCUUUUUUUCGAGCUC -------............((((((((((....))))))))))......((((((((((..(.(((...(((((....)))))....((.........))..))).)..)))))))))). ( -27.00) >consensus _______UGCAGUAAUUCGCUUAGCACUUACUUAAGGCCUAAUAUAU________AGAGCUCUUUAACGCA________UUUGAUAAUCAAAAUUAAGUAAAGAGCUCUCUUUCGAGCUU ........(((((((((........))))))).......................(((((((((((................................))))))))))).......)).. ( -9.41 = -10.97 + 1.56)

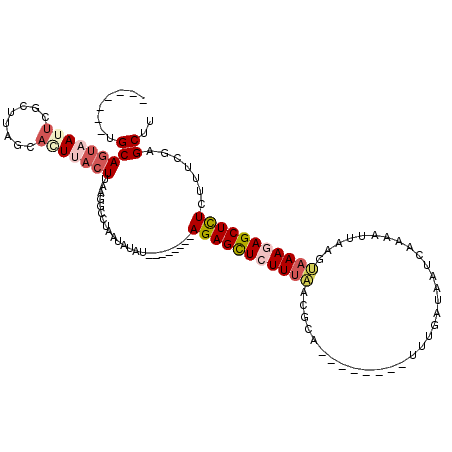
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:17 2006