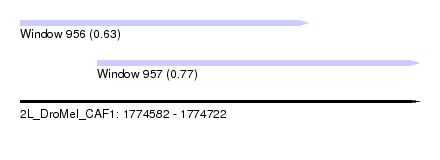
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,774,582 – 1,774,722 |
| Length | 140 |
| Max. P | 0.774678 |

| Location | 1,774,582 – 1,774,683 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 67.38 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -12.72 |
| Energy contribution | -12.76 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1774582 101 + 22407834 ACUUGAAACGGGGUACC-------------CAUAUGCCACCCUCUUCUCACUUCACACAUUUUAAACCAAUUGCGAACUUGGCCCA-----GGCGACAAACACUUAAUUGGUUUAAAGG ....(((..((((((..-------------....)))).))...)))............((((((((((((((.(...(((((...-----.))..)))...).)))))))))))))). ( -20.50) >DroPse_CAF1 36186 113 + 1 --AUGGUAUGUUGUGUUCAACAUGGAGCUCUACCAGUCAUCCCCCUCUGA----CAACAGUUAAAUCCAAUUACGAACUUGGCCCAAACUGAGCAUCAAACAGGUUAUUGAGUUUGAGG --....(((((((....)))))))((((((.(((.((((........)))----)..(((((....((((........))))....)))))...........)))....)))))).... ( -25.70) >DroSec_CAF1 31582 100 + 1 -CUUGAAACGGGGUACC-------------CAUACGCCACCCUCUUCUCAGUUCACACACUUUAAACCAAUUACGAACUUGGCCCA-----GGCGACAAACACUUAAUUGGUUUAAAGG -.((((((.(((((...-------------........))))).)).))))........((((((((((((((.(...(((((...-----.))..)))...).)))))))))))))). ( -23.90) >DroYak_CAF1 40763 100 + 1 ACUUGAAACGGGGUAAC-------------CGUAUGCCACCCU-UUCACACUUCACACACUUUAAACCAAUUACGAACUUGGCCCA-----GGCGACAAGCACUUAAUUGGUUUAAAGG ...(((((.((((((..-------------....)))).)).)-))))...........((((((((((((((.(..((((((...-----.))..))))..).)))))))))))))). ( -31.20) >DroPer_CAF1 35033 113 + 1 --AUGGUAUGGUGUGUUCAACAUGGAGCUCUACCAGUCAUCCCCCUCUGA----CAACAGUUAAAUCCAAUUACGAACUUGGCCCAAACUGAGCAUCAAACAGGUUAUUGAGUUUGAGG --..((..(((((.((((......)))).).)))).....)).((((.((----(..(((((....((((........))))....)))))....((((........))))))).)))) ( -24.60) >consensus _CUUGAAACGGGGUACC_____________CAUAAGCCACCCUCUUCUCA_UUCACACACUUUAAACCAAUUACGAACUUGGCCCA_____GGCGACAAACACUUAAUUGGUUUAAAGG ....((...(((((........................)))))..))............((((((((((((((.....((((((.......)))..))).....)))))))))))))). (-12.72 = -12.76 + 0.04)

| Location | 1,774,609 – 1,774,722 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.87 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -11.36 |
| Energy contribution | -11.56 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1774609 113 + 22407834 CCUCUUCUCACUUCACACAUUUUAAACCAAUUGCGAACUUGGCCCA------GGCGACAAACACUUAAUUGGUUUAAAGGCCUUGACACAAAGUUUUCACUGGCAAUUUGUGAUGG-CUG ..........(.(((((..((((((((((((((.(...(((((...------.))..)))...).))))))))))))))(((.(((..........)))..)))....))))).).-... ( -24.30) >DroPse_CAF1 36224 112 + 1 CCCCCUCUGA----CAACAGUUAAAUCCAAUUACGAACUUGGCCCA-AACUGAGCAUCAAACAGGUUAUUGAGUUUGAGGCCUUGGUAUAAAGUUUUACCUGGCAAUUAG--GUGG-GAU .(((.(((((----...(((.((((......(((.((...((((((-((((.((.(((.....)))..)).)))))).)))))).)))......)))).)))....))))--).))-).. ( -29.20) >DroSec_CAF1 31608 113 + 1 CCUCUUCUCAGUUCACACACUUUAAACCAAUUACGAACUUGGCCCA------GGCGACAAACACUUAAUUGGUUUAAAGGCCUUGACACAAAGUUUUCACUGGCAAUUUGUGAUGG-CUG ........(((((((((..((((((((((((((.(...(((((...------.))..)))...).))))))))))))))(((.(((..........)))..)))....)))))..)-))) ( -29.60) >DroYak_CAF1 40790 112 + 1 CCU-UUCACACUUCACACACUUUAAACCAAUUACGAACUUGGCCCA------GGCGACAAGCACUUAAUUGGUUUAAAGGCCUUGACACAAAGUUUUCACUGGCAAUUUGCGAUGG-CUG ((.-.((.((.........((((((((((((((.(..((((((...------.))..))))..).))))))))))))))(((.(((..........)))..)))....)).)).))-... ( -28.00) >DroAna_CAF1 31831 103 + 1 --------CUGGU-CUACACUUUAAACCAAUUACGAACUUGGCCCACAACCGAGUGACAAUCAUUUAAUUCGUUUAAAGUCCUCGCCACAAAGUAUUGACUUUCAAUUCAAG-------- --------.((((-....(((((((((.(((((...((((((.......))))))(.....)...))))).)))))))))....))))....(.((((.....)))).)...-------- ( -20.00) >DroPer_CAF1 35071 113 + 1 CCCCCUCUGA----CAACAGUUAAAUCCAAUUACGAACUUGGCCCA-AACUGAGCAUCAAACAGGUUAUUGAGUUUGAGGCCUUGGUAUAAAGUUUUACCUGGCACUUAG--AUGGGGAU ((((.(((((----...(((.((((......(((.((...((((((-((((.((.(((.....)))..)).)))))).)))))).)))......)))).)))....))))--).)))).. ( -33.10) >consensus CCUCUUCUCA_UU_ACACACUUUAAACCAAUUACGAACUUGGCCCA______AGCGACAAACACUUAAUUGGUUUAAAGGCCUUGACACAAAGUUUUCACUGGCAAUUUG_GAUGG_CUG ...................((((((((((((((.....(((((..........))..))).....))))))))))))))(((...................)))................ (-11.36 = -11.56 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:12 2006