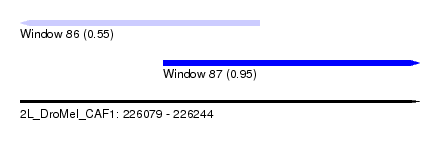
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 226,079 – 226,244 |
| Length | 165 |
| Max. P | 0.948808 |

| Location | 226,079 – 226,178 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 79.24 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -16.10 |
| Energy contribution | -15.13 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
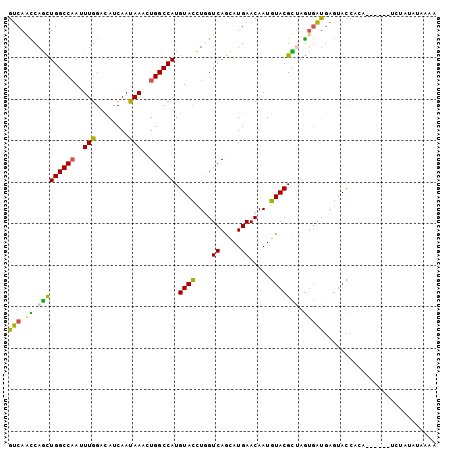
>2L_DroMel_CAF1 226079 99 - 22407834 GUCAACCAGCUGGCCAACUUGGACAUCAAUAAAUUGGCCAUGUACCUGGUCAGCAUGAACAAUGUACGUUAGUGAUGAGUACCUCC----UCUCUAUAUAAAA ((..(((((.(((((((.(((........))).))))))).....)))))..)).........((((.(((....)))))))....----............. ( -22.00) >DroPse_CAF1 22664 96 - 1 GUCAGCCAGUUGGCCAAUUUGGAUAUCAAUAAGCUGGCCAUGUACCUCGUCAGCAUGAACAAUGUACGCUAGUGAUGAGUACCACA-------CAUUAUACUA ((((((..(((((((.....))...)))))..))))))...((((.(((((.((((.....))))((....)))))))))))....-------.......... ( -22.90) >DroEre_CAF1 19460 99 - 1 GUCAACCAGCUGGCCAACUUGGACAUCAAUAAAUUGGCCAUGUACCUGGUCAGCAUGAACAAUGUACGUUAGUGAUGAGUACCUCC----UCUCUAAAUAAAA ((..(((((.(((((((.(((........))).))))))).....)))))..))..............((((.((.(((...))).----)).))))...... ( -23.10) >DroWil_CAF1 25006 103 - 1 GUCAGUCAAUUGGCCAAUUUGGAUAUCAACAAACUGGCCAUGUAUUUGGUCAGUAUGAACAAUGUACGUAAAAAGCCCAUAUCACAAUCUACUUUAUAGCAAA (((((....))))).....(((((.....((.((((((((......)))))))).))....(((..(.......)..)))......)))))............ ( -18.40) >DroAna_CAF1 24790 96 - 1 GUCAAUCAGCUGGCCCAGUUGGACAUCAACAAGCUGGCCAUGUACCUGGUCAGCAUGAACAAUGUACGCGAGUGAUGAGUACCCA-------UAUAUGCCAUA ..........((((.....(((...(((.((.((((((((......)))))))).)).......(((....))).)))....)))-------.....)))).. ( -27.40) >DroPer_CAF1 22922 96 - 1 GUCAGCCAGUUGGCCAAUUUGGAUAUCAAUAAGCUGGCCAUGUACCUCGUCAGCAUGAACAAUGUACGCUAGUGAUGAGUACCACA-------CAUUAUACUA ((((((..(((((((.....))...)))))..))))))...((((.(((((.((((.....))))((....)))))))))))....-------.......... ( -22.90) >consensus GUCAACCAGCUGGCCAAUUUGGACAUCAAUAAACUGGCCAUGUACCUGGUCAGCAUGAACAAUGUACGCUAGUGAUGAGUACCACA______UCUAUAUAAAA (((.((.(((((((((..(((........)))..)))))).((((....((.....)).....))))))).)))))........................... (-16.10 = -15.13 + -0.97)

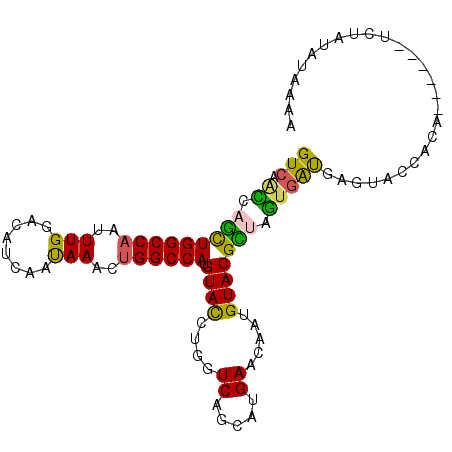
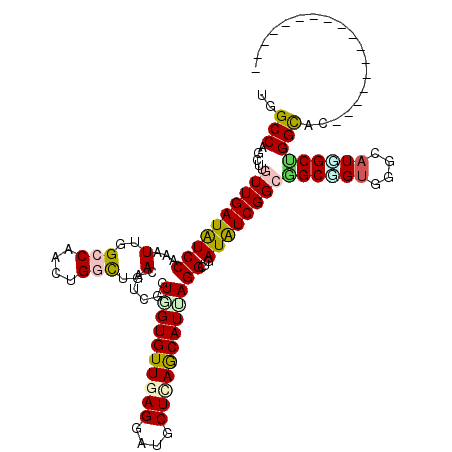
| Location | 226,138 – 226,244 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.19 |
| Mean single sequence MFE | -42.92 |
| Consensus MFE | -29.44 |
| Energy contribution | -29.62 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 226138 106 + 22407834 UGGCCAAUUUAUUGAUGUCCAAGUUGGCCAGCUGGUUGACAUCUGUGGUGUUGAGUAUGCUGAGCAUUAGGCUUAUGUCGGCGGCGGUUGGCAUAGCUGGCAUGAC-------------- ((((((((((..........))))))))))((((.(((((((..((((((((.((....)).))))))..))..))))))))))).((..((...))..)).....-------------- ( -38.70) >DroVir_CAF1 22740 103 + 1 UGGCCAGUUUGUUGAUAUCCAAGUUGGCCAAUUGGCUGACAUCGGUUGUGUUGAGGAUGCUCAGCAUAAGGCUGAUAUCGGCGGCUGUGGGCAUGGCUGGCAC----------------- ..(((((((((((.(((.((..((..(((....)))..))(((((((((((((((....)))))))))..))))))......)).))).)))).)))))))..----------------- ( -46.60) >DroGri_CAF1 21885 103 + 1 UGGCCAGCUUGUUGAUAUCCAAAUUGGCCAAUUGGCUGACAUCUGUUGUGUUGAGGAUGCUCAGCAUCAGGCUUAUGUCGGCAGCGGUGGGCAUUGCAGGCAC----------------- (((((((.(((........))).)))))))....((((((((..(((((((((((....))))))))..)))..)))))))).(((((....)))))......----------------- ( -39.70) >DroWil_CAF1 25069 106 + 1 UGGCCAGUUUGUUGAUAUCCAAAUUGGCCAAUUGACUGACAUCGGUUGUGUUGAGGAUGCUUAGCAUUAGGCUAAUAUCGGCAGCAGUUGGCAUGGCUGGUACUCC-------------- (((((((((((........)))))))))))..........((((((..(((..(.(.((((((((.....)))).....)))).)..)..)))..)))))).....-------------- ( -37.70) >DroAna_CAF1 24846 120 + 1 UGGCCAGCUUGUUGAUGUCCAACUGGGCCAGCUGAUUGACGUCGGUGGUGUUGAGAAUGCUGAGCAUCAGGCUGAUGUCGGCGGCCGAGGGCAUGGCCGGCAUUCCCGAUCCGGUCACAG (((((.(.(((..((((((...((.((((.(((....(((((((((((((((.((....)).))))))..)))))))))))))))).))(((...)))))))))..))).).)))))... ( -51.40) >DroPer_CAF1 22978 103 + 1 UGGCCAGCUUAUUGAUAUCCAAAUUGGCCAACUGGCUGACAUCGGUGGUGUUGAGGAUGCUCAACAUUAGGCUGAUAUCGGCGGCGGUGGGCAUCGCCGGCAC----------------- (((((((....(((.....))).)))))))....(((((.(((((((((((((((....)))))))))..)))))).)))))((((((....)))))).....----------------- ( -43.40) >consensus UGGCCAGCUUGUUGAUAUCCAAAUUGGCCAACUGGCUGACAUCGGUGGUGUUGAGGAUGCUCAGCAUUAGGCUGAUAUCGGCGGCGGUGGGCAUGGCUGGCAC_________________ ..(((.....((((((((((...(..(((....)))..)......((((((((((....))))))))))))...))))))))((((((....)))))))))................... (-29.44 = -29.62 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:24:34 2006