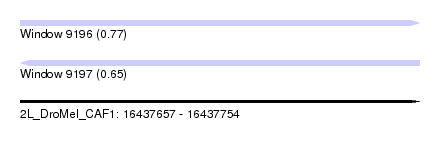
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,437,657 – 16,437,754 |
| Length | 97 |
| Max. P | 0.765488 |

| Location | 16,437,657 – 16,437,754 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 76.94 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -13.20 |
| Energy contribution | -13.53 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
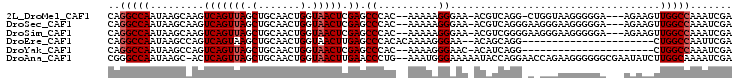
>2L_DroMel_CAF1 16437657 97 + 22407834 CAGGCCAAUAAGCAAGUCAGUUAGCUGCAACUGGUAACUCGAGCCCAC--AAAAAGGGAA-ACGUCAGG-CUGGUAAGGGGGA---AGAAGUUGGCCAAAUCGA ..(((((((..(((.((......))))).((..((....((..(((..--.....)))..-.))....)-)..))........---....)))))))....... ( -26.80) >DroSec_CAF1 62065 98 + 1 CAGGCCAAUAAGCAAGUCAGUUAGCUGCAACUGGUAACUCGAGCCCAC--AAAAAGGGAA-ACGUCAGGGAAGGGAAGGGGGA---AGAAGUUGGCCAAAUCGA ..(((((((.......((((((......))))))...(((...(((.(--....(.(...-.).)...)...)))..)))...---....)))))))....... ( -25.00) >DroSim_CAF1 62413 98 + 1 CAGGCCAAUAAGCAAGUCAGUUAGCUGCAACUGGUAACUCGAGCCCAC--AAAAAGGGAA-ACGUCGGGGAAGGGAAGGGGGA---AGAAGUUGGCCAAAUCGA ..(((((((.......((((((......))))))...(((((.(((..--.....)))..-...)))))..............---....)))))))....... ( -25.00) >DroEre_CAF1 59783 80 + 1 CAGGCCAAUAAGCCAGUCAGUAAGCUGCAACUGGUAACUUGAGCCCACACAAAAGGGAA--ACAGCAGG----------------------CUGGCCAAUUCGA ..(((((....(((((((((....)))..))))))..((((..(((........)))..--....))))----------------------.)))))....... ( -26.10) >DroYak_CAF1 63294 79 + 1 CAGGCCAAUAAGCCAGUCAGUUAGCUGCAACUGGUAACUCGAGCCCAC--AAAAGGGAAC-ACAUCAGG----------------------CUGGCCAAAUCGA ..(((((....(((((((((....)))..)))))).....(..(((..--....)))..)-........----------------------.)))))....... ( -24.60) >DroAna_CAF1 56450 101 + 1 CGGGCCAAUAAGC-ACUCAGUUAGCUGCAACUGGUAACUUGAACCCUG--AAAUGGGAAAAAUACCAGGAACCAGAAGGGGGGCGAAUAUCUUGGCAAAAUCGA ...(((((...((-.(((......(((...((((((..((...(((..--....))).))..))))))....)))....))))).......)))))........ ( -25.30) >consensus CAGGCCAAUAAGCAAGUCAGUUAGCUGCAACUGGUAACUCGAGCCCAC__AAAAAGGAAA_ACAUCAGG_A_GGGAAGGGGGA___AGAAGUUGGCCAAAUCGA ..(((((.........(((((((.(.......).))))).)).((..........))...................................)))))....... (-13.20 = -13.53 + 0.33)
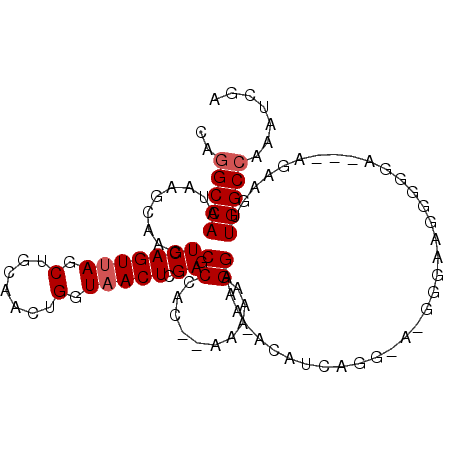

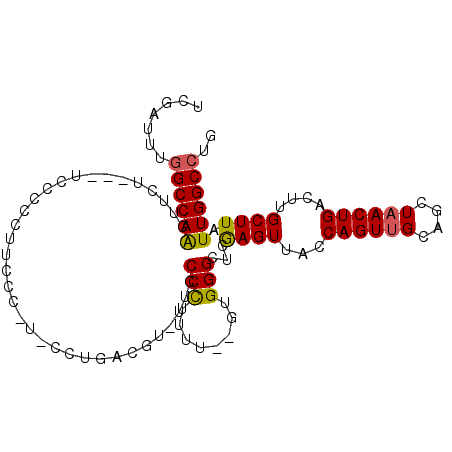
| Location | 16,437,657 – 16,437,754 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 76.94 |
| Mean single sequence MFE | -22.14 |
| Consensus MFE | -17.48 |
| Energy contribution | -17.12 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16437657 97 - 22407834 UCGAUUUGGCCAACUUCU---UCCCCCUUACCAG-CCUGACGU-UUCCCUUUUU--GUGGGCUCGAGUUACCAGUUGCAGCUAACUGACUUGCUUAUUGGCCUG .......((((((.....---...........((-(((.(((.-.........)--)))))))(((((...((((((....)))))))))))....)))))).. ( -22.80) >DroSec_CAF1 62065 98 - 1 UCGAUUUGGCCAACUUCU---UCCCCCUUCCCUUCCCUGACGU-UUCCCUUUUU--GUGGGCUCGAGUUACCAGUUGCAGCUAACUGACUUGCUUAUUGGCCUG .......((((((.....---......................-..(((.....--..)))..(((((...((((((....)))))))))))....)))))).. ( -19.80) >DroSim_CAF1 62413 98 - 1 UCGAUUUGGCCAACUUCU---UCCCCCUUCCCUUCCCCGACGU-UUCCCUUUUU--GUGGGCUCGAGUUACCAGUUGCAGCUAACUGACUUGCUUAUUGGCCUG .......((((((.....---..............(((.(((.-.........)--)))))..(((((...((((((....)))))))))))....)))))).. ( -20.40) >DroEre_CAF1 59783 80 - 1 UCGAAUUGGCCAG----------------------CCUGCUGU--UUCCCUUUUGUGUGGGCUCAAGUUACCAGUUGCAGCUUACUGACUGGCUUAUUGGCCUG .......((((((----------------------((..(...--...........)..)))........(((((..(((....)))))))).....))))).. ( -23.84) >DroYak_CAF1 63294 79 - 1 UCGAUUUGGCCAG----------------------CCUGAUGU-GUUCCCUUUU--GUGGGCUCGAGUUACCAGUUGCAGCUAACUGACUGGCUUAUUGGCCUG .......((((((----------------------((((((.(-(..(((....--..)))..)).)))).((((((....))))))...)))....))))).. ( -23.20) >DroAna_CAF1 56450 101 - 1 UCGAUUUUGCCAAGAUAUUCGCCCCCCUUCUGGUUCCUGGUAUUUUUCCCAUUU--CAGGGUUCAAGUUACCAGUUGCAGCUAACUGAGU-GCUUAUUGGCCCG ........(((((..(((((............((..((((((.(((.(((....--..)))...))).))))))..))((....))))))-)....)))))... ( -22.80) >consensus UCGAUUUGGCCAACUUCU___UCCCCCUUCCC_U_CCUGACGU_UUCCCCUUUU__GUGGGCUCGAGUUACCAGUUGCAGCUAACUGACUUGCUUAUUGGCCUG .......((((((..................................(((........)))...((((...((((((....))))))....)))).)))))).. (-17.48 = -17.12 + -0.36)

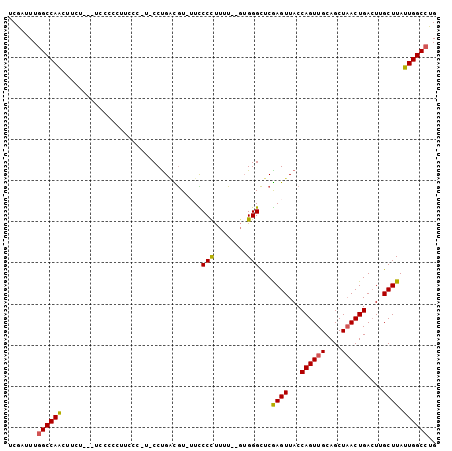
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:12 2006