
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,431,294 – 16,431,457 |
| Length | 163 |
| Max. P | 0.844921 |

| Location | 16,431,294 – 16,431,389 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -19.54 |
| Consensus MFE | -11.70 |
| Energy contribution | -11.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16431294 95 + 22407834 -----CCGCACCUU-CCCAAACACCCUACCCUGCACCACAGACACC---GCAUGGCCGACAAAAUAAUAUCAUUUGACGAUCAAAGUAAUUAUUUUGCUUUGGU -----.........-.(((((.......((.(((............---))).))..(.(((((((((....((((.....))))...))))))))))))))). ( -14.70) >DroPse_CAF1 33106 102 + 1 --AGCCUAAGCGUCCCCCCGCCCCCCUGCCCCUCGGCACAGACACCGCCGCAUGGCCGACAAAAUAAUAUCAUUUGACGAUCAAAGUAAUUAUUUUGCUUUGGU --.(((.(((((((............((((....))))........(((....))).)))((((((((....((((.....))))...)))))))))))).))) ( -23.60) >DroEre_CAF1 50543 99 + 1 GCACCCCGCACCC--CCCAAACACCCUGCCCUGCGCCACAGACACC---GCAUGGCCGACAAAAUAAUAUCAUUUGACGAUCAAAGUAAUUAUUUUGCUUUGGU ((.....))....--.(((((......(((.((((..........)---))).))).(.(((((((((....((((.....))))...))))))))))))))). ( -20.80) >DroYak_CAF1 56967 99 + 1 ACACCCCGCACCC--CCCAAACACCAUGCCCUGCACCACAGACACC---GCAUGGCCGACAAAAUAAUAUCAUUUGACGAUCAAAGUAAUUAUUUUGCUUUGGU .............--.(((((..((((((..(((......).))..---))))))..(.(((((((((....((((.....))))...))))))))))))))). ( -20.10) >DroAna_CAF1 50496 94 + 1 --ACCCCACUCCGU-----CAGCCCCUUUUCUGCACCACAGACACC---GCAUGGCCGACAAAAUAAUAUCAUUUGACGAUCAAAGUAAUUAUUUUGCUUUGGU --..........((-----(.(((.....((((.....))))....---....))).)))...................((((((((((.....)))))))))) ( -18.72) >DroPer_CAF1 31560 89 + 1 ---------------CGCCCACCCCCUGCCCCUCGGCACAGACACCGCCGCAUGGCCGACAAAAUAAUAUCAUUUGACGAUCAAAGUAAUUAUUUUGCUUUGGU ---------------...(((......(((...((((.........))))...))).(.(((((((((....((((.....))))...))))))))))..))). ( -19.30) >consensus __ACCCCGCACCC__CCCAAACACCCUGCCCUGCACCACAGACACC___GCAUGGCCGACAAAAUAAUAUCAUUUGACGAUCAAAGUAAUUAUUUUGCUUUGGU ......................................................((((((((((((((....((((.....))))...)))))))))..))))) (-11.70 = -11.70 + 0.00)

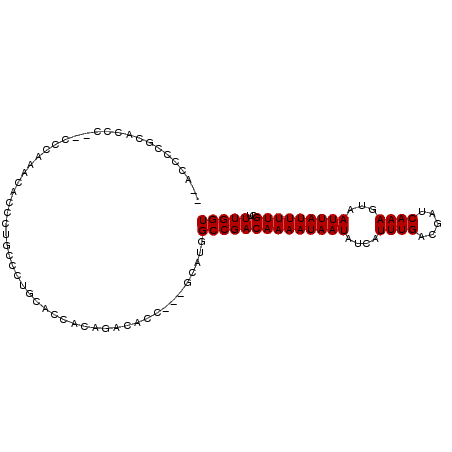
| Location | 16,431,324 – 16,431,429 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.49 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -12.22 |
| Energy contribution | -12.38 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.844921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16431324 105 + 22407834 --------CACAGACA---CCGCAUGGC-CGACAAAAUAAUAUCAUUUGACGAUCAAAGUAAUUAUUUUGCUUUGGUCUACAGCAUAAUUGCGGGUUCUACUCCGACUGCGGGGUUG --------...(((..---(((((..((-.(.((((.........))))..(((((((((((.....)))))))))))..).)).....)))))..)))((((((....)))))).. ( -31.90) >DroVir_CAF1 29184 95 + 1 --------GCCGUACA---CC-CAUUGC-CGACAAAAUAAUAUCAUUUG-CGAUCAAAGUAAUUAUUUUGCUUUGGUCUACAGAAUAAUUACGGCUUCUGCGCCGC------GGC-- --------(((((...---..-....((-((.((((.........))))-.(((((((((((.....))))))))))).............)))).........))------)))-- ( -25.97) >DroGri_CAF1 33221 95 + 1 --------GCCGUAUA---CC-CAUUGU-CGACAAAAUAAUAUCAUUUU-CGAUCAAAGUAAUUAUUUUGCUUUGGUCUACAGCAUAAUUACGGCUUCUACGGUGC------GGC-- --------(((((((.---..-....((-((..(((((......)))))-.(((((((((((.....))))))))))).............)))).......))))------)))-- ( -26.94) >DroWil_CAF1 59520 102 + 1 AACUCACUCAUACAUAUGGCCACAUGGC-CGACAAAAUAAUAUCAUUUGACGAUCAAAGUAAUUAUUUUGCUUUGGUCUACAGCAUAAUUGCGGGUUCUACUA-------------- (((((...........(((((....)))-))....................(((((((((((.....)))))))))))....(((....))))))))......-------------- ( -27.10) >DroMoj_CAF1 26021 96 + 1 --------UCUGUACU---CC-CAUUGCCCGACAAAAUAAUAUCAUUUG-CGAUCAAAGUAAUUAUUUCGCUUUGGUCUACAGAAUAAUUACGGCUUCAACGGCGU------GGU-- --------((((((..---..-........(.((((.........))))-)((((((((..(.....)..))))))))))))))...((((((.(......).)))------)))-- ( -20.50) >DroAna_CAF1 50525 105 + 1 --------CACAGACA---CCGCAUGGC-CGACAAAAUAAUAUCAUUUGACGAUCAAAGUAAUUAUUUUGCUUUGGUCUACAGCAUAAUUGCGGGUUCUACUCGGAACAAGGGGUUG --------....(((.---(((((..((-.(.((((.........))))..(((((((((((.....)))))))))))..).)).....)))))(((((....))))).....))). ( -27.60) >consensus ________CACAUACA___CC_CAUGGC_CGACAAAAUAAUAUCAUUUG_CGAUCAAAGUAAUUAUUUUGCUUUGGUCUACAGCAUAAUUACGGCUUCUACGCCGA______GGU__ ...................................................(((((((((((.....)))))))))))....................................... (-12.22 = -12.38 + 0.17)
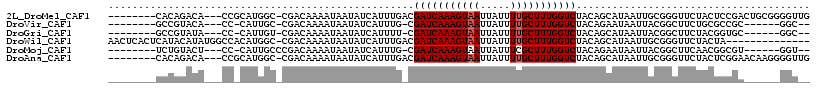

| Location | 16,431,349 – 16,431,457 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.23 |
| Mean single sequence MFE | -34.54 |
| Consensus MFE | -19.04 |
| Energy contribution | -20.00 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.600672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16431349 108 + 22407834 UAAUAUCAUUUGACGAUCAAAGUAAUUAUUUUGCUUUGGUCUACAGCAUAAUUGCGGGUUCUACUCCGACUGCGGGGUUGGCCAAGGGUUAAGGCUGG--GGGGAGC----------GAG .....((.(((..((((((((((((.....)))))))))))..((((.(((((...((((..((((((....)))))).))))...)))))..)))).--)..))).----------)). ( -34.30) >DroPse_CAF1 33168 106 + 1 UAAUAUCAUUUGACGAUCAAAGUAAUUAUUUUGCUUUGGUCUACAGCAUAAUUGUGGGUUCUACGCCGACUA-GGGGUUGUCCAGGGCUC-G-GCUCGGCUCGGCC-----------CGG ..............(((((((((((.....))))))))))).(((((......((.(((.....))).))..-...)))))((.(((((.-(-((...))).))))-----------))) ( -34.30) >DroYak_CAF1 57026 119 + 1 UAAUAUCAUUUGACGAUCAAAGUAAUUAUUUUGCUUUGGUCUACAGCAUAAUUGCGGGUUCUACUCCGACUGCGGGGUUGGCCUAGGGUUAAGGCCGG-AGGGGUGCUGAGGAGAAAGAG .....((.(((..((((((((((((.....)))))))))))..((((((..((.((((((..((((((....)))))).)))))..(((....)))).-))..)))))).)..))).)). ( -42.00) >DroAna_CAF1 50550 107 + 1 UAAUAUCAUUUGACGAUCAAAGUAAUUAUUUUGCUUUGGUCUACAGCAUAAUUGCGGGUUCUACUCGGAACAAGGGGUUGGACCGGGGAUAGGG--GG-AGGGUAGA----------GAG ...((((.(((..((((((((((((.....))))))))))).........(((.(.(((((.((((........)))).))))).).))).)..--))-).))))..----------... ( -30.30) >DroPer_CAF1 31609 101 + 1 UAAUAUCAUUUGACGAUCAAAGUAAUUAUUUUGCUUUGGUCUACAGCAUAAUUGUGGGUUCUACGCCGACUA-GGGGUUGUCCAGGGCUC-G-GCUCGGCCCG----------------G ..............(((((((((((.....)))))))))))....(((....)))(((((....(((((((.-.((.....))..)).))-)-))..))))).----------------. ( -31.80) >consensus UAAUAUCAUUUGACGAUCAAAGUAAUUAUUUUGCUUUGGUCUACAGCAUAAUUGCGGGUUCUACUCCGACUA_GGGGUUGGCCAGGGGUUAGGGCUGG_AGGGGAG___________GAG ..............(((((((((((.....)))))))))))....(((....))).((((..(((((......))))).))))..................................... (-19.04 = -20.00 + 0.96)

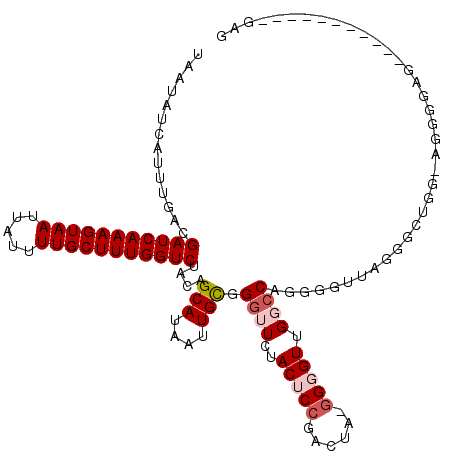
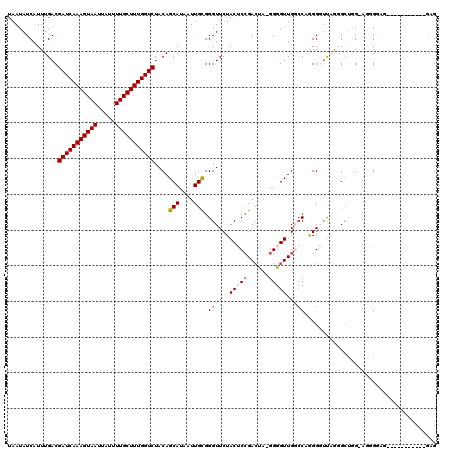
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:07 2006