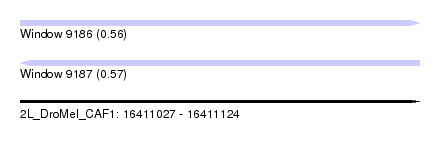
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,411,027 – 16,411,124 |
| Length | 97 |
| Max. P | 0.569340 |

| Location | 16,411,027 – 16,411,124 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 80.18 |
| Mean single sequence MFE | -21.08 |
| Consensus MFE | -10.42 |
| Energy contribution | -9.79 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16411027 97 + 22407834 CUUUAGCGCAUU--UC-UU-UUUCUAAUUGUAAGCGGAUAUUUAUUCUCCAUCAUUUCCGCUCAUUUGAAUUUUUUCGCCCCACUCGGACGGGAAAAGGAA ...........(--((-((-(((((.......((((((..................))))))..............((((......)).)))))))))))) ( -18.57) >DroSec_CAF1 35347 98 + 1 CUUUAGCGCAUU--UCCUUUUUUCUAAUUGUAAGCGGAUAUUUAUUCUCCAUCAUUUCCGCUCAUUUGAAUUUUUUCGCCCCACUUGGGCG-GAAAAGGAA ...........(--(((....(((.((.((..((((((..................)))))))).))))).((((((((((.....)))))-))))))))) ( -24.67) >DroSim_CAF1 34978 97 + 1 CUUUAGCGCAUU--UCCUU-UUUCUAAUUGUAAGCGGAUAUUUAUUCUCCAUCAUUUCCGCUCAUUUGAAUUUUUUCGCCCCACUUGGGCG-GAAAAGGAA ...........(--(((((-((((........((((((..................)))))).....(((....)))((((.....)))))-))))))))) ( -25.57) >DroEre_CAF1 31002 97 + 1 CUUUAGCUCGAUACUC-UU-UGUCUAAUUGUAAGCGGAUAUUUGUUCUCCAUCUUUUCCGCUCAUUUGAAUUUUCUCGCCCCACUUGGGUG-GGA-AGGAA ..((((..(((.....-.)-)).)))).....((((((....((.....)).....))))))........(((((((((((.....)))))-)))-))).. ( -24.10) >DroYak_CAF1 36393 96 + 1 CUUUAGCUCAUU--UC-UU-UGUCUAAUUGUAAGCGGAUAUUUAUUCUCCAUCAUUUCCGCUCAUUUGAAUUUUUUCGGCCCUCUUGGAUG-GAAAAGGAA ...........(--((-((-(.((((......((((((..................))))))...(((((....))))).((....)).))-)).)))))) ( -15.47) >DroAna_CAF1 30383 78 + 1 --------------------UUUCUUAUUGUCGGCGGAUAUUUAUUUCCCAUAAUUUCCGCUCAUUUUAAUUUUCCCU-CGUACUCCC--GGGGAUGGCUA --------------------.........(((((((((...((((.....))))..))))))...........((((.-((......)--))))).))).. ( -18.10) >consensus CUUUAGCGCAUU__UC_UU_UUUCUAAUUGUAAGCGGAUAUUUAUUCUCCAUCAUUUCCGCUCAUUUGAAUUUUUUCGCCCCACUUGGGCG_GAAAAGGAA ................................((((((..................))))))........((((((((.((.....)).)).))))))... (-10.42 = -9.79 + -0.64)

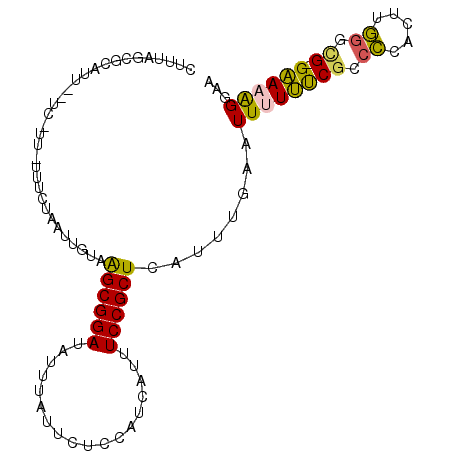
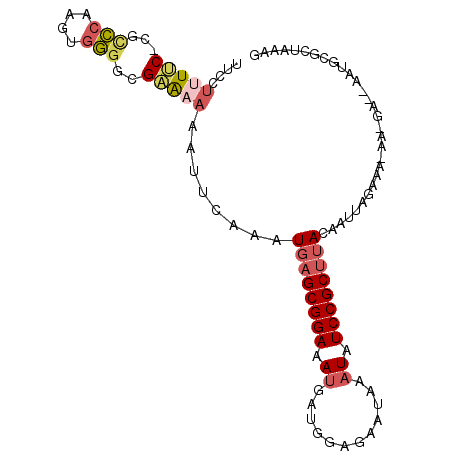
| Location | 16,411,027 – 16,411,124 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.18 |
| Mean single sequence MFE | -21.62 |
| Consensus MFE | -10.94 |
| Energy contribution | -11.55 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16411027 97 - 22407834 UUCCUUUUCCCGUCCGAGUGGGGCGAAAAAAUUCAAAUGAGCGGAAAUGAUGGAGAAUAAAUAUCCGCUUACAAUUAGAAA-AA-GA--AAUGCGCUAAAG (((.(((((.(((((.....)))))............((((((((.((............)).))))))))......))))-).-))--)........... ( -21.40) >DroSec_CAF1 35347 98 - 1 UUCCUUUUC-CGCCCAAGUGGGGCGAAAAAAUUCAAAUGAGCGGAAAUGAUGGAGAAUAAAUAUCCGCUUACAAUUAGAAAAAAGGA--AAUGCGCUAAAG ((((((((.-(((((.....)))))............((((((((.((............)).)))))))).........)))))))--)........... ( -26.90) >DroSim_CAF1 34978 97 - 1 UUCCUUUUC-CGCCCAAGUGGGGCGAAAAAAUUCAAAUGAGCGGAAAUGAUGGAGAAUAAAUAUCCGCUUACAAUUAGAAA-AAGGA--AAUGCGCUAAAG ((((((((.-(((((.....)))))............((((((((.((............)).))))))))........))-)))))--)........... ( -26.90) >DroEre_CAF1 31002 97 - 1 UUCCU-UCC-CACCCAAGUGGGGCGAGAAAAUUCAAAUGAGCGGAAAAGAUGGAGAACAAAUAUCCGCUUACAAUUAGACA-AA-GAGUAUCGAGCUAAAG .....-.((-(((....)))))((..((..((((...((((((((.....((.....))....))))))))(.....)...-..-)))).))..))..... ( -21.50) >DroYak_CAF1 36393 96 - 1 UUCCUUUUC-CAUCCAAGAGGGCCGAAAAAAUUCAAAUGAGCGGAAAUGAUGGAGAAUAAAUAUCCGCUUACAAUUAGACA-AA-GA--AAUGAGCUAAAG .(((((((.-.....)))))))...............((((((((.((............)).))))))))...((((...-.(-..--..)...)))).. ( -16.10) >DroAna_CAF1 30383 78 - 1 UAGCCAUCCCC--GGGAGUACG-AGGGAAAAUUAAAAUGAGCGGAAAUUAUGGGAAAUAAAUAUCCGCCGACAAUAAGAAA-------------------- ......(((((--(......))-.)))).........((.(((((..((((.....))))...)))))))...........-------------------- ( -16.90) >consensus UUCCUUUUC_CGCCCAAGUGGGGCGAAAAAAUUCAAAUGAGCGGAAAUGAUGGAGAAUAAAUAUCCGCUUACAAUUAGAAA_AA_GA__AAUGCGCUAAAG ....(((((...(((....)))..)))))........((((((((.((............)).)))))))).............................. (-10.94 = -11.55 + 0.61)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:02 2006