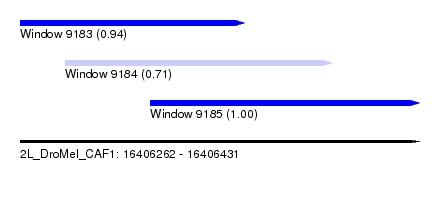
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,406,262 – 16,406,431 |
| Length | 169 |
| Max. P | 0.999886 |

| Location | 16,406,262 – 16,406,357 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 88.91 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -23.47 |
| Energy contribution | -24.50 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
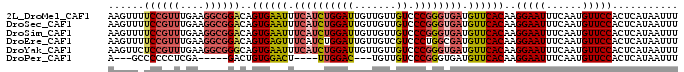
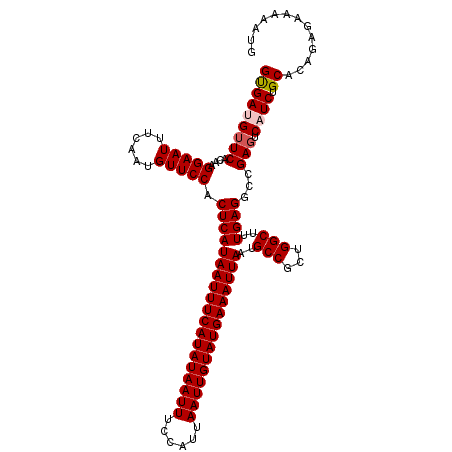
>2L_DroMel_CAF1 16406262 95 + 22407834 AAGUUUUCCGUUUGAAGGCGGACAGUGAAUUUCAUCUGGAUUGUUGUUGUCCCGGGUGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUU ......((((((....))))))..((((((.((((((((((.......)).)))))))).))))))..(((((......)))))........... ( -29.90) >DroSec_CAF1 30669 95 + 1 AAGUUUUCCGUUUGAAGGCGGACAGUGAAUUUCAUCUGGAUUGUUGUUGUCCCGGGUGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUU ......((((((....))))))..((((((.((((((((((.......)).)))))))).))))))..(((((......)))))........... ( -29.90) >DroSim_CAF1 30218 95 + 1 AAGUUUUCCGUUUGAAGGCGGACAGUGAAUUUCAUCUGGAUUGUUGUUGUCCCGGGUGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUU ......((((((....))))))..((((((.((((((((((.......)).)))))))).))))))..(((((......)))))........... ( -29.90) >DroEre_CAF1 26265 95 + 1 AAGUUUUCCGUUUGAAGGCGGACAGUGAGUUUCAUCUGGAUUGUUGUCGUCCCUGGCGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUU ......((((((....))))))..((((((......((((..(((((((....)))))))..)).)).(((((......)))))))))))..... ( -24.30) >DroYak_CAF1 31505 95 + 1 AAGUUCUCCGUUUGAAGGCGGGCAGUGAAUUUCAUCUGGAUUGUUGUUGUCCCGGGUGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUU ..((((.((.......)).)))).((((((.((((((((((.......)).)))))))).))))))..(((((......)))))........... ( -29.60) >DroPer_CAF1 4717 80 + 1 A---GCCCCCCUCGA-----GACUGUGGACU----UUGGAC---UGUUGUCCCGGGUGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUU .---.........((-----(..(((((((.----((((((---....)))).....)).))))))).(((((......))))).)))....... ( -20.60) >consensus AAGUUUUCCGUUUGAAGGCGGACAGUGAAUUUCAUCUGGAUUGUUGUUGUCCCGGGUGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUU ......((((((....))))))..((((((.((((((((((.......)).)))))))).))))))..(((((......)))))........... (-23.47 = -24.50 + 1.03)
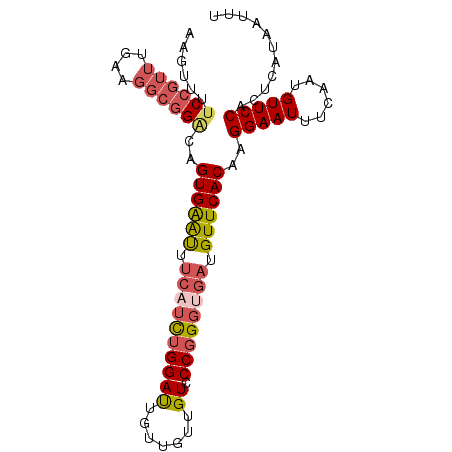
| Location | 16,406,281 – 16,406,394 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 86.10 |
| Mean single sequence MFE | -32.95 |
| Consensus MFE | -21.87 |
| Energy contribution | -24.40 |
| Covariance contribution | 2.53 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16406281 113 + 22407834 GGAC----AGUGAAUUUCAUCUGGAUUGUUGUUGUCCCGGGUGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGC- ((.(----(((((((.((((((((((.......)).)))))))).))))))..(((((......))))).....(((((((((((((((......))))))))))))))).))))..- ( -39.20) >DroPse_CAF1 4777 99 + 1 -GAC----UGUGG-----------AC---UGUUGUCCCGGGUGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGCAUGAAAUGAAUGCCCCC -...----.(.((-----------((---....)))))(((.(..((((....(((((......))))).......(((((((.(((((......))))).)))))))))))..)))) ( -25.30) >DroEre_CAF1 26284 113 + 1 GGAC----AGUGAGUUUCAUCUGGAUUGUUGUCGUCCCUGGCGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGC- ((.(----(((((((......((((..(((((((....)))))))..)).)).(((((......)))))))))))((((((((((((((......))))))))))))))..))))..- ( -32.50) >DroYak_CAF1 31524 113 + 1 GGGC----AGUGAAUUUCAUCUGGAUUGUUGUUGUCCCGGGUGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAGGCCGC- .(((----.((((((.((((((((((.......)).)))))))).))))))..(((((......))))).....(((((((((((((((......)))))))))))))))..)))..- ( -42.00) >DroAna_CAF1 26004 117 + 1 GAGCUAGUAGACAAUUUUGUCUGGUUUGUUGUUGUCCCGGGUGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGC- (((.....(((((....)))))((..(((((..((.((..(((.....)))..)).))..)))))..)).))).(((((((((((((((......))))))))))))))).......- ( -33.60) >DroPer_CAF1 4729 105 + 1 -GAC----UGUGGACU----UUGGAC---UGUUGUCCCGGGUGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGCAUGAAAUGAAUGCCCC- -...----.(((((((----..((((---....))))..))(((.((((.....)))).)))....))))).(((.(((((((.(((((......))))).)))))))..)))....- ( -25.10) >consensus GGAC____AGUGAAUUUCAUCUGGAUUGUUGUUGUCCCGGGUGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGC_ .........((((((.((.(((((............))))).)).))))))..(((((......))))).....(((((((((((((((......)))))))))))))))........ (-21.87 = -24.40 + 2.53)
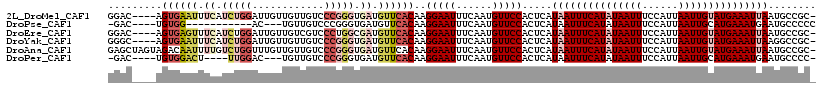
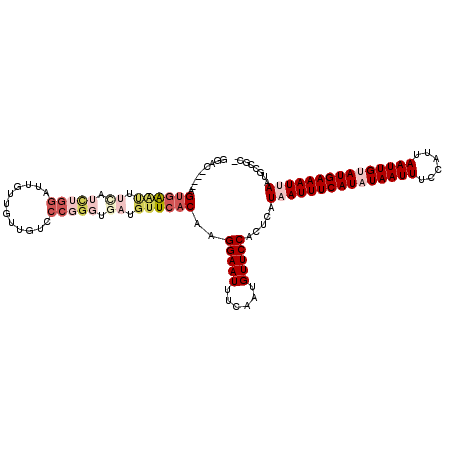
| Location | 16,406,317 – 16,406,431 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 96.37 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -29.72 |
| Energy contribution | -30.08 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.74 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.39 |
| SVM RNA-class probability | 0.999886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16406317 114 + 22407834 GUGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGCUGGCUUUGAGGCCGAGUCAUCUGCACAGAGAAAAAUG (((.....)))..(((((......))))).(((.(((((((((((((((......))))))))))))))).(((.(.((((((.......)))))).).)))..)))....... ( -32.50) >DroSec_CAF1 30724 114 + 1 GUGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGCUGGCUUUGAGGCCGAGUCUUCUGCACAGAGAAAAAUG (((.....)))..(((((......))))).(((.(((((((((((((((......))))))))))))))).(((.(((((((....)))).))).....)))..)))....... ( -32.30) >DroSim_CAF1 30273 114 + 1 GUGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGCUGGCUUUGAGGCCGAGUCAUCUGCACAGAGAAAAAUG (((.....)))..(((((......))))).(((.(((((((((((((((......))))))))))))))).(((.(.((((((.......)))))).).)))..)))....... ( -32.50) >DroEre_CAF1 26320 114 + 1 GCGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGCUGGCUUUGAGGCCGAGUCAUCUGCACAGCGAAAAAUG ((((((.......(((((......)))))((((.(((((((((((((((......)))))))))))))))........((((....)))))))))))).))............. ( -33.50) >DroYak_CAF1 31560 114 + 1 GUGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAGGCCGCUGGCUUUGAGGCCGAGUCAUCUGCACAGAGAAAAACG ((((((.......(((((......)))))((((.(((((((((((((((......))))))))))))))).((((.(.......).)))))))))))).))............. ( -34.20) >DroAna_CAF1 26044 114 + 1 GUGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGCUGGCUUUGAGACAGACUCUUCUGCACAGAACAAAAAA ....(((((....(((((......))))).(((((((((((((((((((......)))))))))))))))..(((...)))..)))).((((....))))....)))))..... ( -31.90) >consensus GUGAUGUUCACAAGGAAUUUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGCUGGCUUUGAGGCCGAGUCAUCUGCACAGAGAAAAAUG (((((((((....(((((......))))).(((((((((((((((((((......)))))))))))))))..(((...)))..))))...))).)))).))............. (-29.72 = -30.08 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:01 2006