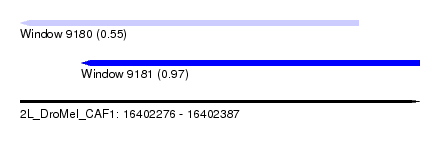
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,402,276 – 16,402,387 |
| Length | 111 |
| Max. P | 0.974836 |

| Location | 16,402,276 – 16,402,370 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -21.42 |
| Consensus MFE | -11.93 |
| Energy contribution | -12.67 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
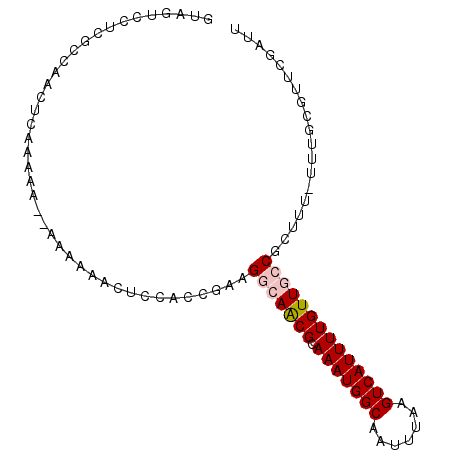
>2L_DroMel_CAF1 16402276 94 - 22407834 GUAGUCUUCGCCAACUUAAAAAAAAAAAAACUCCACCGAAGGCAACGCAAAUGGCAAUUUAAGUCAUUUUGUUUGCCGUUU-UUUGCGUUCGAUU ...(((((((..........................)))))))((((((((((((((..((((....)))).)))))))..-.)))))))..... ( -22.27) >DroSec_CAF1 26727 90 - 1 GUAGUCCUCGCCAACUUCAAAA--AAAUAACUCCACCGAAGGCAACGCAAAUGGCAAUUUAAGUCAUUUUGUUUGCG--UU-UUUGCGUUCGAUU ......................--............((((.((((((((((((((.......))))).....)))))--).-..))).))))... ( -18.20) >DroEre_CAF1 22712 82 - 1 ------------AAAUCAAAAAAAAAAAAAACCCACCGAAGGCAGCGCAAAUGGCAAUUUAAGUCAUUUUGUUGCCGCUUU-UUUGAGUGCGAUU ------------.((((.......................(((((((.(((((((.......))))))))))))))((((.-...))))..)))) ( -20.50) >DroYak_CAF1 27528 91 - 1 GAAGGCCUCGCCAAGUCAAA----AAAUAACUCCACCGAAGGCAGCGCAAAUGGCAAUUUAAGUCAUUUUGUUGCCGCUUUUUUUGCGUUCGAUU ...(((...)))........----............(((((((((((.(((((((.......))))))))))))))((.......)).))))... ( -24.70) >consensus GUAGUCCUCGCCAACUCAAAAA__AAAAAACUCCACCGAAGGCAACGCAAAUGGCAAUUUAAGUCAUUUUGUUGCCGCUUU_UUUGCGUUCGAUU ........................................(((((((.(((((((.......))))))))))))))................... (-11.93 = -12.67 + 0.75)

| Location | 16,402,293 – 16,402,387 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 76.90 |
| Mean single sequence MFE | -16.00 |
| Consensus MFE | -11.93 |
| Energy contribution | -12.67 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16402293 94 - 22407834 CACUCCCCAAAAACC-CCGUAGUCUUCGCCAACUUAAAAAAAAAAAAACUCCACCGAAGGCAACGCAAAUGGCAAUUUAAGUCAUUUUGUUUGCC ..........((((.-.(((.(((((((..........................))))))).))).(((((((.......))))))).))))... ( -14.37) >DroSec_CAF1 26742 93 - 1 CACUCCCCAAAAACCCCCGUAGUCCUCGCCAACUUCAAAA--AAAUAACUCCACCGAAGGCAACGCAAAUGGCAAUUUAAGUCAUUUUGUUUGCG ................................((((....--.............))))((((.(((((((((.......)))).))))))))). ( -11.93) >DroEre_CAF1 22729 79 - 1 GCCAACUUAAAAAA----------------AAAUCAAAAAAAAAAAAAACCCACCGAAGGCAGCGCAAAUGGCAAUUUAAGUCAUUUUGUUGCCG ..............----------------............................(((((((.(((((((.......)))))))))))))). ( -17.30) >DroYak_CAF1 27546 91 - 1 CACUCCCCGAAAAACCCCGAAGGCCUCGCCAAGUCAAA----AAAUAACUCCACCGAAGGCAGCGCAAAUGGCAAUUUAAGUCAUUUUGUUGCCG ..................((.(((...)))...))...----................(((((((.(((((((.......)))))))))))))). ( -20.40) >consensus CACUCCCCAAAAAAC_CCGUAGUCCUCGCCAACUCAAAAA__AAAAAACUCCACCGAAGGCAACGCAAAUGGCAAUUUAAGUCAUUUUGUUGCCG ..........................................................(((((((.(((((((.......)))))))))))))). (-11.93 = -12.67 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:57 2006