| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,400,228 – 16,400,333 |
| Length | 105 |
| Max. P | 0.739035 |

| Location | 16,400,228 – 16,400,333 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.78 |
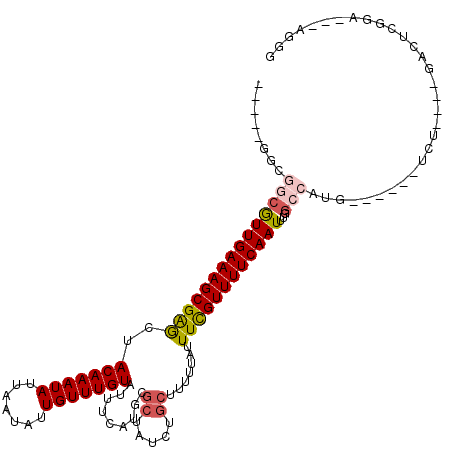
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -16.95 |
| Energy contribution | -17.45 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.739035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
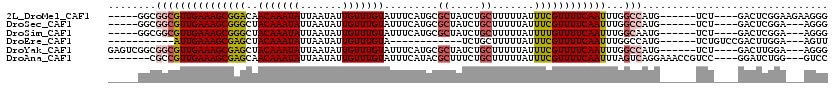
>2L_DroMel_CAF1 16400228 105 + 22407834 -----GGCGGCGUUGAAAGCGGACAACAAAUAUUAAUAUUGUUUGUAUUUCAUGCGCUAUCUGCUUUUUAUUUCGUUUUCAAUUUGGCCAUG------UCU----GACUCGGAAGAAGGG -----(((.(.((((((((((((..(((((((.......))))))).........((.....)).......)))))))))))).).)))...------.((----...((....)).)). ( -28.20) >DroSec_CAF1 24680 102 + 1 -----GGCGGCGUUGAAAGCGGGCUACAAAUAUUAAUAUUGUUUGUAUUUCAUGCGCUAUCUGCUUUUUAUUUCGUUUUCAAUUUGGCCAUG------UCU----GACUCGGA---AGGG -----(((.(.((((((((((((.((((((((.......))))))))........((.....)).......)))))))))))).).)))...------...----..(((...---.))) ( -29.60) >DroSim_CAF1 23423 102 + 1 -----GGCGGCGUUGAAAGCGGGCUACAAAUAUUAAUAUUGUUUGUAUUUCAUGCGCUAUCUGCUUUUUAUUUUGUUUUCAAUUUGGCAAUG------UCU----GACUCGGA---AGGG -----.((.(.((((((((((((.((((((((.......)))))))).)))....((.....))..........))))))))).).))....------...----..(((...---.))) ( -25.50) >DroEre_CAF1 20751 88 + 1 -----------AUUGAAAGCGAGCUACAAAUAUUAAUAUUGUUUGUA------------UCUGCUUUUUAUUUCGUUUUCAAUUUGGCCAUG------UCUGUCCGACUUGGA---AGUU -----------(((((((((((((((((((((.......))))))))------------...))........)))))))))))....(((.(------((.....))).))).---.... ( -21.30) >DroYak_CAF1 25437 107 + 1 GAGUCGGCGGCGUUGAAAGCGAGCUACAAAUAUUAAUAUUGUUUGUAUUUCAUGCGCUAUCUGCUUUUUAUUUCGUUUUCAAUUUGGCCAUG------UCU----GACUUGGA---AGGG (((((((((((((((((((((((.((((((((.......))))))))........((.....)).......))))))))))))...)))..)------.))----)))))...---.... ( -30.70) >DroAna_CAF1 19634 106 + 1 -------CGCCGUUGAAAGCGAGCAACAAAUAUUAAUAUUGUUUGUAUUUCAUACGCUUUCUGCUUUUUAUUUCGUUUUCAAUUUAGUCAGGAAACCGUCC----GGAUCUGG---GUCC -------.(((((((((((((((..(((((((.......))))))).........((.....)).......)))))))))))).(((((.(((.....)))----.)).))))---)).. ( -25.30) >consensus _____GGCGGCGUUGAAAGCGAGCUACAAAUAUUAAUAUUGUUUGUAUUUCAUGCGCUAUCUGCUUUUUAUUUCGUUUUCAAUUUGGCCAUG______UCU____GACUCGGA___AGGG ........(((((((((((((((..(((((((.......))))))).........((.....)).......))))))))))))...)))............................... (-16.95 = -17.45 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:52 2006