
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,399,674 – 16,399,925 |
| Length | 251 |
| Max. P | 0.875381 |

| Location | 16,399,674 – 16,399,794 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.26 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -19.63 |
| Energy contribution | -21.87 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16399674 120 + 22407834 UUCUCCACUUACACAAAAAUUUACAUUUGAUUGCUGUUCAAUCAACUCUCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGUUGUUGGCUCUGUGGGACC ..........................(((((((.....)))))))......(((....)))((((((((((.(((((..(.(((...........))).)..)))))))))))))))... ( -30.50) >DroSec_CAF1 24129 118 + 1 UUCUCCACUUACA--GAAAUUUACAUUUGAUUGCUGUUCAAUCAACUCUCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGCUGUUGGCUCUGUGGGACC ...........((--(..........(((((((.....))))))).........)))....((((((((((.(((((.((.(((...........)))..)))))))))))))))))... ( -31.21) >DroSim_CAF1 22874 118 + 1 UUCUCCACUUACA--GAAAUUUACAUUUGAUUGCUGUUCAAUCAACUCUCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGUUGUUGGCUCUGUGGGACC ...........((--(..........(((((((.....))))))).........)))....((((((((((.(((((..(.(((...........))).)..)))))))))))))))... ( -31.11) >DroEre_CAF1 20263 92 + 1 UUCUCCACUUAUG--GCAAUUUACAUUUGAUUGCUGUUCAAUCAACUC--------------CCACAGAGCUCAACAUGUGAUAUCCCUGACAUUUAUGAGUUCCUGA------------ ..........(((--((((((.......)))))))))...........--------------...(((((((((..((((.(......).))))...)))))).))).------------ ( -17.80) >DroYak_CAF1 24875 118 + 1 UUCUCUAUUCACA--GAAAUUUACAUUUGAUUGCUGUUCAAUCAACUCUCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGAUGUUGGCUCUGUGGGACC ........((.((--(..........(((((((.....))))))).........)))..))((((((((((.((((((.(.(((...........))).).))))))))))))))))... ( -34.81) >DroAna_CAF1 19021 118 + 1 GGCUCCUUUUCGA--GCAAUUUACAUUCGAUUACUGUCCUAUCUACUUUGAUCACUGACAACCCACAGAGUUCAACAUGUGAUAUCCUUGACAGUUAUGAGUUGUUGGCCCUGUGGGUCC .((((......))--)).................((((..(((......)))....))))((((((((.(..(((((..(.(((...........))).)..)))))..))))))))).. ( -29.40) >consensus UUCUCCACUUACA__GAAAUUUACAUUUGAUUGCUGUUCAAUCAACUCUCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGUUGUUGGCUCUGUGGGACC ..........................(((((((.....)))))))................(((((((((.((((((..(.(((...........))).)..)))))))))))))))... (-19.63 = -21.87 + 2.24)

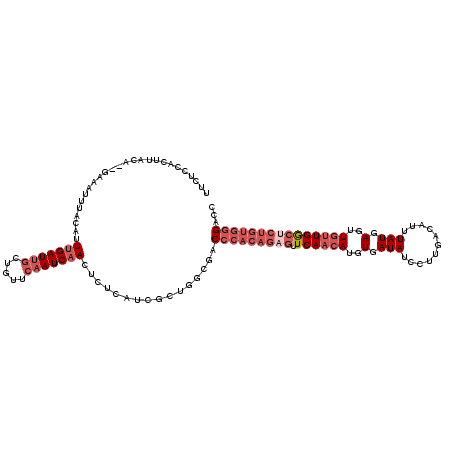
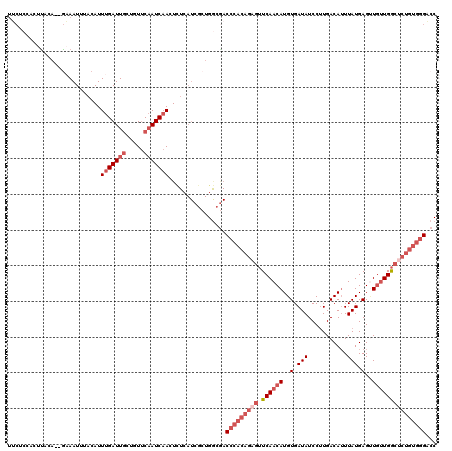
| Location | 16,399,714 – 16,399,827 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.81 |
| Mean single sequence MFE | -29.86 |
| Consensus MFE | -24.36 |
| Energy contribution | -25.24 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16399714 113 + 22407834 AUCAACUCUCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGUUGUUGGCUCUGUGGGACCCCCGUUCCCCUA-------UACAAAAUAACAUGUUAGAUC ..............((((((.((((((((((.(((((..(.(((...........))).)..)))))))))))))))......((.......-------.)).........))))))... ( -28.20) >DroSec_CAF1 24167 113 + 1 AUCAACUCUCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGCUGUUGGCUCUGUGGGACCCCCGCUCCCCUA-------UACAAAGUAACAUGUUAGAUC .............((.((.(.((((((((((.(((((.((.(((...........)))..))))))))))))))))).).)).))....(((-------.(((........))))))... ( -29.90) >DroSim_CAF1 22912 113 + 1 AUCAACUCUCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGUUGUUGGCUCUGUGGGACCCCCGCUCCCCUA-------UACAAAGUAACAUGUUAGAUC .............((.((.(.((((((((((.(((((..(.(((...........))).)..))))))))))))))).).)).))....(((-------.(((........))))))... ( -29.80) >DroYak_CAF1 24913 113 + 1 AUCAACUCUCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGAUGUUGGCUCUGUGGGACCCCCGCUCCCCUA-------UACAACGUAACAUGUUAGAUC .............((.((.(.((((((((((.((((((.(.(((...........))).).)))))))))))))))).).)).))....(((-------.(((........))))))... ( -33.30) >DroAna_CAF1 19059 120 + 1 AUCUACUUUGAUCACUGACAACCCACAGAGUUCAACAUGUGAUAUCCUUGACAGUUAUGAGUUGUUGGCCCUGUGGGUCCCCCACUCUACUAUGUCCUCUGUAAGGAAGCAUGUUAUAUC .........((.(.(((........))).).))(((((((....(((((((((.....((((.(..(((((...)))))..).)))).....))))......))))).)))))))..... ( -28.10) >consensus AUCAACUCUCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGUUGUUGGCUCUGUGGGACCCCCGCUCCCCUA_______UACAAAGUAACAUGUUAGAUC .............((.((.(.((((((((((.(((((..(.(((...........))).)..))))))))))))))).).)).))................................... (-24.36 = -25.24 + 0.88)


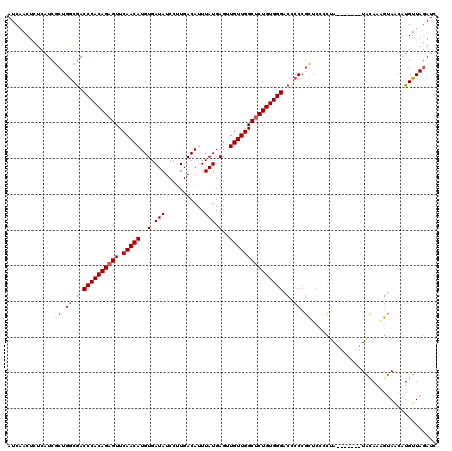
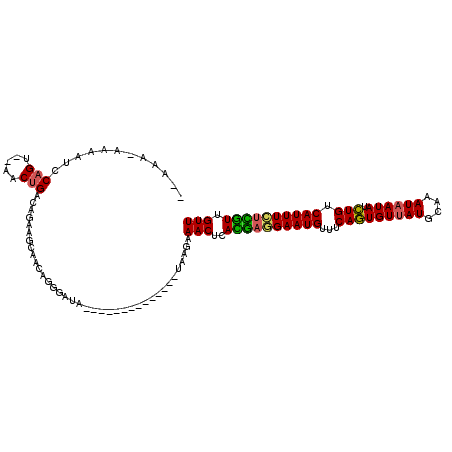
| Location | 16,399,827 – 16,399,925 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.09 |
| Mean single sequence MFE | -19.74 |
| Consensus MFE | -16.30 |
| Energy contribution | -15.74 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16399827 98 - 22407834 --AAAAGAAAUCCAGU--AACUGACAGAAGCAACAGGGAUA--------------UAAGAAACUCACGAGGAAUGUUUCAGUGUUAUGCAAAUAAUAUCUGUCAUUUCUCGUUGUU --.......((((.((--..((......))..))..)))).--------------.....(((..((((((((((...(((((((((....)))))).))).)))))))))).))) ( -23.30) >DroSec_CAF1 24280 97 - 1 --AAA-AAAAUCCAGU--AACUGACAGAAGCAACAGGGAUA--------------UAAGAAACUCACGAGGAAUGUUCCAGUGUUAUGCAAAUAAUAUCUGUCAUUUCUCGUUGUU --...-...((((.((--..((......))..))..)))).--------------.....(((..((((((((((...(((((((((....)))))).))).)))))))))).))) ( -22.60) >DroSim_CAF1 23025 95 - 1 ----A-AAAAUCCAGU--AACUGACAGAAGCAACAGGGAUA--------------UAAGAAACUCACGAGGAAUGUUUCAGUGUUAUGCAAAUAAUAUCUGUCAUUUCUCGUUGUU ----.-...((((.((--..((......))..))..)))).--------------.....(((..((((((((((...(((((((((....)))))).))).)))))))))).))) ( -23.30) >DroYak_CAF1 25026 94 - 1 ------AAAAUCCAGU--AACCGACAGAAGCAACAGGAAUA--------------CAAAAAACUCAUGAAGAAUGUUUCAGUGUUAUGCAAAUAAUAUCUGUCAUUUUGUGUUGUU ------..........--..........((((((((.....--------------).............((((((...(((((((((....)))))).))).)))))).))))))) ( -14.10) >DroAna_CAF1 19179 116 - 1 GAAAAAAACACACAGUCUCACUGACAGAAGCAACAGGGAAGUUACUGAACAACAAAAAAAAACUCAUAAGGAAUGUUUCAAUGUUAUGCAAAUCAUAUUUGACAUUUGUUAUUGUU ..............(((.....)))...((((((((........)))...................(((.(((((((..(((((.((....)).))))).))))))).)))))))) ( -15.40) >consensus __AAA_AAAAUCCAGU__AACUGACAGAAGCAACAGGGAUA______________UAAGAAACUCACGAGGAAUGUUUCAGUGUUAUGCAAAUAAUAUCUGUCAUUUCUCGUUGUU ............(((.....))).....................................(((..((((((((((...(((((((((....)))))).))).)))))))))).))) (-16.30 = -15.74 + -0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:51 2006