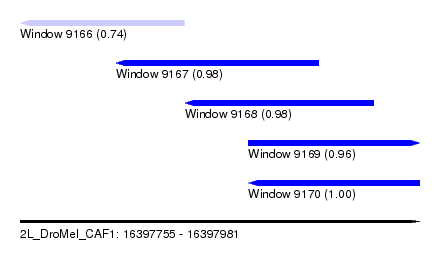
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,397,755 – 16,397,981 |
| Length | 226 |
| Max. P | 0.999150 |

| Location | 16,397,755 – 16,397,848 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.66 |
| Mean single sequence MFE | -21.06 |
| Consensus MFE | -13.76 |
| Energy contribution | -14.98 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
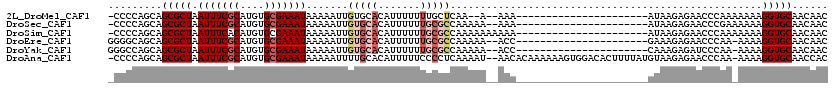
>2L_DroMel_CAF1 16397755 93 - 22407834 -CCCCAGCAGCGCUAAUUUCGCAUGUGCGAAAUAAAAAUUGUGCACAUUUUUUUGCUCAA--A--AAA----------------------AUAAGAGAACCCAAAAAAAGGUGCAACAAC -....(((...))).(((((((....))))))).....((((((((.(((((((((((..--.--...----------------------....)))....)))))))).)))).)))). ( -21.90) >DroSec_CAF1 22200 95 - 1 -CCCCAGCAGCGCUAAUUUCGCAUGUGCGAAAUAAAAAUUGUGCACAUUUUUUGCGCCAAAAA--AAA----------------------AUAAGAGAACCCGAAAAAAGGUGCAACAAC -.....(((((((..(((((((....)))))))(((((.((....)).)))))))))......--...----------------------..........((.......)))))...... ( -19.60) >DroSim_CAF1 20955 97 - 1 -CCCCAGCAGCGCUAAUUUCACAUGUGCGAAAUAAAAAUUGUGCACAUUUUUUGCGCCAAAAAAAAAA----------------------AUAAGAGAACCCAAAAAAAGGUGCAACAAC -.....((.((((.........(((((((.(((....))).))))))).....))))...........----------------------........(((........)))))...... ( -16.64) >DroEre_CAF1 18528 95 - 1 GGGGCAGCAGCGCUAAUUUCGCAUGUGCGAAAUAAAAAUUGUGCACAUUUUUUGCGCCAAAAA--ACC----------------------GAAAGAGAACCCAA-AAAAGGUGCAACAAC (..(((.(.((((..(((((((....)))))))(((((.((....)).)))))))))......--..(----------------------....).........-....).)))..)... ( -23.90) >DroYak_CAF1 22851 95 - 1 GGGCCAGCAGCGCUAAUUUCGCAUGUGCGAAAUAAAAAUUGUGCACAUUUUUUGCGCCAAAAA--ACC----------------------CAAAGAGAUCCCAA-AAAAGGUGCAACAAC (((......((((..(((((((....)))))))(((((.((....)).)))))))))......--.))----------------------).........((..-....))......... ( -20.12) >DroAna_CAF1 17433 116 - 1 -CCCCAGCAGCGCUAAUUUCGCAUGUGCGAAAUAAAAAUUUUGCACAUUUUUCCCCUCAAAAU--AACACAAAAAAGUGGACACUUUUAUGUAAGAGAACCCAA-AAAAGGUGCAACCAC -........(((((........(((((((((((....)))))))))))...............--...(((.((((((....)))))).)))............-....)))))...... ( -24.20) >consensus _CCCCAGCAGCGCUAAUUUCGCAUGUGCGAAAUAAAAAUUGUGCACAUUUUUUGCGCCAAAAA__AAA______________________AUAAGAGAACCCAA_AAAAGGUGCAACAAC .........(((((.(((((((....))))))).......(((((.......)))))....................................................)))))...... (-13.76 = -14.98 + 1.22)
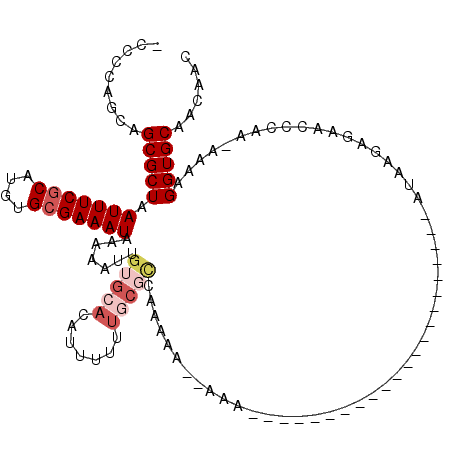
| Location | 16,397,809 – 16,397,924 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.94 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -24.27 |
| Energy contribution | -24.02 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16397809 115 - 22407834 GACAUUGUUCAUAGUCAUAACAUGACGGAAAAAUGUCAAUUUAAAUAUUUGCAAAUAUUUUAACAAGGCAUGUGCG-----CCCCAGCAGCGCUAAUUUCGCAUGUGCGAAAUAAAAAUU ((((((.(((...(((((...))))).))).)))))).....(((((((....))))))).......(((((((((-----....(((...))).....)))))))))............ ( -27.60) >DroSec_CAF1 22256 115 - 1 GACAUUGUUCAUAGUCAUAACGUGACGGAAAAAUGUCAAUUUAAAUAUUUGCAAAUAUUUUAACAAGGCAUGUGCG-----CCCCAGCAGCGCUAAUUUCGCAUGUGCGAAAUAAAAAUU ((((((.(((...(((((...))))).))).)))))).....(((((((....))))))).......(((((((((-----....(((...))).....)))))))))............ ( -27.90) >DroSim_CAF1 21013 115 - 1 GACAUUGUUCAUAGUCAUAACAUGACGGAAAAAUGUCAAUUUAAAUAUUUGCAAAUAUUUUAACAAGGCAUGUGCG-----CCCCAGCAGCGCUAAUUUCACAUGUGCGAAAUAAAAAUU ((((((.(((...(((((...))))).))).))))))..........(((((.((....))......(((((((((-----(.......)))).......)))))))))))......... ( -24.51) >DroEre_CAF1 18583 117 - 1 GACAUUGUUCAUAGUCAUAACAUGACGGAAAAAUGUCAAUUUAAAUAUUUGCAAAUAUUUUAACAAGGCAUGUGCU---UGGGGCAGCAGCGCUAAUUUCGCAUGUGCGAAAUAAAAAUU ((((((.(((...(((((...))))).))).)))))).....(((((((....))))))).......(((((((((---(((.((....)).))))....))))))))............ ( -29.90) >DroYak_CAF1 22906 120 - 1 GACAUUGUUCAUAGUCAUAACAUGACGGAAAAAUGUCAAUUUAAAUAUUUGCAAAUAUUUUAACAAGGCAUGUGCUGCCGGGGCCAGCAGCGCUAAUUUCGCAUGUGCGAAAUAAAAAUU ((((((.(((...(((((...))))).))).)))))).....(((((((....)))))))...........(((((((.(....).)))))))..(((((((....)))))))....... ( -34.60) >DroAna_CAF1 17510 114 - 1 GACAUUGUUCAUAGUCAUAACAUGACGGAAAAAUGUCAAUUUAAAUAUUUGCAAAUAUUUUAACAAGGUAUGCGA------CCCCAGCAGCGCUAAUUUCGCAUGUGCGAAAUAAAAAUU ((((((.(((...(((((...))))).))).)))))).....(((((((....)))))))......(((.(((..------.....)))..))).(((((((....)))))))....... ( -26.40) >consensus GACAUUGUUCAUAGUCAUAACAUGACGGAAAAAUGUCAAUUUAAAUAUUUGCAAAUAUUUUAACAAGGCAUGUGCG_____CCCCAGCAGCGCUAAUUUCGCAUGUGCGAAAUAAAAAUU ((((((.(((...(((((...))))).))).)))))).....(((((((....)))))))......(((.(((.............)))..))).(((((((....)))))))....... (-24.27 = -24.02 + -0.25)

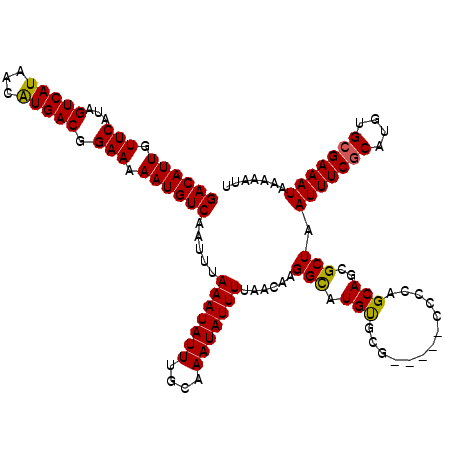
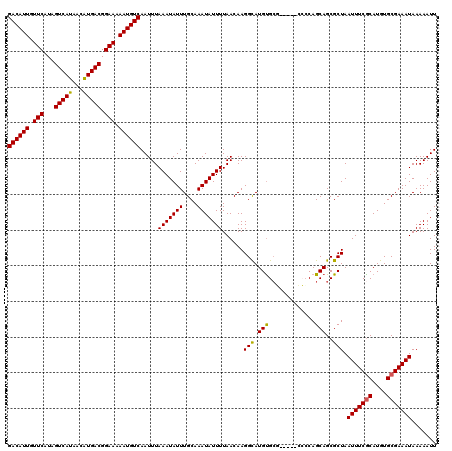
| Location | 16,397,848 – 16,397,955 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.44 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -24.46 |
| Energy contribution | -24.21 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16397848 107 - 22407834 CCUUUGGGCCUUCAAG--------GCAUCCAGCUA-AAUUGACAUUGUUCAUAGUCAUAACAUGACGGAAAAAUGUCAAUUUAAAUAUUUGCAAAUAUUUUAACAAGGCAUGUGCG---- ....((((((.....)--------))..)))((((-((((((((((.(((...(((((...))))).))).)))))))))))(((((((....)))))))......))).......---- ( -27.60) >DroSec_CAF1 22295 107 - 1 CCUUUGGGCCUUCAAG--------GCAUCCAGCUA-AAUUGACAUUGUUCAUAGUCAUAACGUGACGGAAAAAUGUCAAUUUAAAUAUUUGCAAAUAUUUUAACAAGGCAUGUGCG---- ....((((((.....)--------))..)))((((-((((((((((.(((...(((((...))))).))).)))))))))))(((((((....)))))))......))).......---- ( -27.90) >DroSim_CAF1 21052 107 - 1 CCUUUGGGCCUUCAAG--------GCAUCCAGCUA-AAUUGACAUUGUUCAUAGUCAUAACAUGACGGAAAAAUGUCAAUUUAAAUAUUUGCAAAUAUUUUAACAAGGCAUGUGCG---- ....((((((.....)--------))..)))((((-((((((((((.(((...(((((...))))).))).)))))))))))(((((((....)))))))......))).......---- ( -27.60) >DroEre_CAF1 18623 107 - 1 CCUUU-GGCCUUCGAG--------GCAUCCAGCUA-AAUUGACAUUGUUCAUAGUCAUAACAUGACGGAAAAAUGUCAAUUUAAAUAUUUGCAAAUAUUUUAACAAGGCAUGUGCU---U .((((-((((.....)--------)).......((-((((((((((.(((...(((((...))))).))).))))))))))))....................)))))........---. ( -27.10) >DroYak_CAF1 22946 111 - 1 CCUUUGGGCCUUCAAG--------GCAUCCAGCUA-AAUUGACAUUGUUCAUAGUCAUAACAUGACGGAAAAAUGUCAAUUUAAAUAUUUGCAAAUAUUUUAACAAGGCAUGUGCUGCCG ...(((((((.....)--------))..)))).((-((((((((((.(((...(((((...))))).))).)))))))))))).......................((((.....)))). ( -30.60) >DroAna_CAF1 17549 115 - 1 ACCUCGAGCCUUGAGGCGAGUUACCCGCCCAGCAAAAAUUGACAUUGUUCAUAGUCAUAACAUGACGGAAAAAUGUCAAUUUAAAUAUUUGCAAAUAUUUUAACAAGGUAUGCGA----- ...(((.((((((.((((.......))))..(((((((((((((((.(((...(((((...))))).))).))))))))))......)))))...........))))))...)))----- ( -35.90) >consensus CCUUUGGGCCUUCAAG________GCAUCCAGCUA_AAUUGACAUUGUUCAUAGUCAUAACAUGACGGAAAAAUGUCAAUUUAAAUAUUUGCAAAUAUUUUAACAAGGCAUGUGCG____ .......(((((............(((.........((((((((((.(((...(((((...))))).))).))))))))))........)))............)))))........... (-24.46 = -24.21 + -0.25)

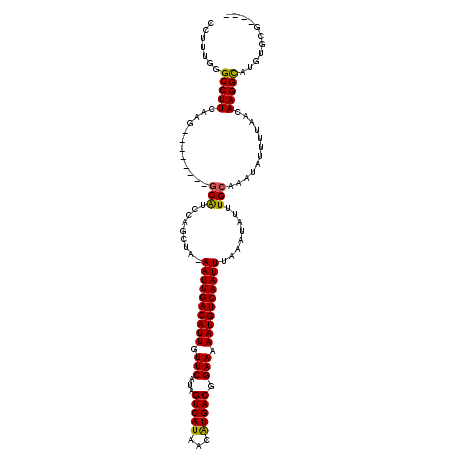

| Location | 16,397,884 – 16,397,981 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.73 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -13.16 |
| Energy contribution | -12.52 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16397884 97 + 22407834 AUUGACAUUUUUCCGUCAUGUUAUGACUAUGAACAAUGUCAAUU-UAGCUGGAUGC--------CUUGAAGGCCCAAAGGUUUUUUUGUCCUGGACCCUCUCC--UUU (((((((((.(((.(((((...)))))...))).))))))))).-.....(((((.--------...(((((((....))))))).))))).(((.....)))--... ( -22.90) >DroSec_CAF1 22331 95 + 1 AUUGACAUUUUUCCGUCACGUUAUGACUAUGAACAAUGUCAAUU-UAGCUGGAUGC--------CUUGAAGGCCCAAAGGUUU--UUAUCCUGCUCCCUUUCC--UCU (((((((((.(((.((((.....))))...))).))))))))).-.(((.((((..--------...(((((((....)))))--)))))).)))........--... ( -23.70) >DroSim_CAF1 21088 95 + 1 AUUGACAUUUUUCCGUCAUGUUAUGACUAUGAACAAUGUCAAUU-UAGCUGGAUGC--------CUUGAAGGCCCAAAGGUUU--UUAUCCUGCUCCCUUUCC--GCU (((((((((.(((.(((((...)))))...))).))))))))).-.(((.(((.((--------..((((((((....)))))--)))....))......)))--))) ( -24.30) >DroEre_CAF1 18660 96 + 1 AUUGACAUUUUUCCGUCAUGUUAUGACUAUGAACAAUGUCAAUU-UAGCUGGAUGC--------CUCGAAGGCC-AAAGGUUUCCUUUUUUUGGCUCCUCUCC--UUU (((((((((.(((.(((((...)))))...))).))))))))).-.....(((.((--------(..(((((((-...))...)))))....)))))).....--... ( -23.80) >DroAna_CAF1 17584 107 + 1 AUUGACAUUUUUCCGUCAUGUUAUGACUAUGAACAAUGUCAAUUUUUGCUGGGCGGGUAACUCGCCUCAAGGCUCGAGGUG-UUCUCUUUUUUUUUUUUCUCCUCGUU (((((((((.(((.(((((...)))))...))).)))))))))....((((((((((...)))))))...))).(((((.(-.................).))))).. ( -30.13) >consensus AUUGACAUUUUUCCGUCAUGUUAUGACUAUGAACAAUGUCAAUU_UAGCUGGAUGC________CUUGAAGGCCCAAAGGUUU__UUAUCCUGCUCCCUCUCC__UUU (((((((((.(((.((((.....))))...))).)))))))))..........................(((.....(((.........)))....)))......... (-13.16 = -12.52 + -0.64)


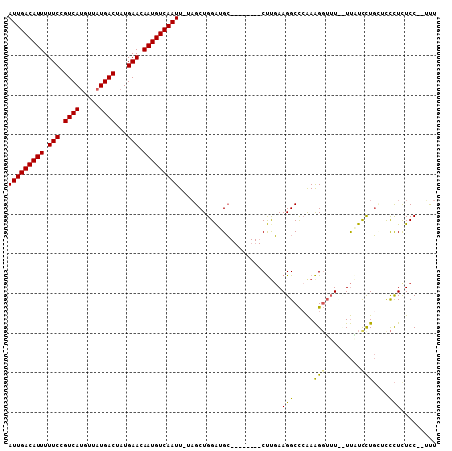
| Location | 16,397,884 – 16,397,981 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.73 |
| Mean single sequence MFE | -29.19 |
| Consensus MFE | -17.90 |
| Energy contribution | -18.14 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -4.03 |
| Structure conservation index | 0.61 |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.999150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16397884 97 - 22407834 AAA--GGAGAGGGUCCAGGACAAAAAAACCUUUGGGCCUUCAAG--------GCAUCCAGCUA-AAUUGACAUUGUUCAUAGUCAUAACAUGACGGAAAAAUGUCAAU ...--...(((((((((((...........)))))))))))..(--------((.....))).-.(((((((((.(((...(((((...))))).))).))))))))) ( -31.90) >DroSec_CAF1 22331 95 - 1 AGA--GGAAAGGGAGCAGGAUAA--AAACCUUUGGGCCUUCAAG--------GCAUCCAGCUA-AAUUGACAUUGUUCAUAGUCAUAACGUGACGGAAAAAUGUCAAU ...--........(((.((((..--...((...))(((.....)--------)))))).))).-.(((((((((.(((...(((((...))))).))).))))))))) ( -27.20) >DroSim_CAF1 21088 95 - 1 AGC--GGAAAGGGAGCAGGAUAA--AAACCUUUGGGCCUUCAAG--------GCAUCCAGCUA-AAUUGACAUUGUUCAUAGUCAUAACAUGACGGAAAAAUGUCAAU (((--(((((((...........--...))))...(((.....)--------)).))).))).-.(((((((((.(((...(((((...))))).))).))))))))) ( -27.54) >DroEre_CAF1 18660 96 - 1 AAA--GGAGAGGAGCCAAAAAAAGGAAACCUUU-GGCCUUCGAG--------GCAUCCAGCUA-AAUUGACAUUGUUCAUAGUCAUAACAUGACGGAAAAAUGUCAAU ...--...((((.((((((....(....).)))-)))))))..(--------((.....))).-.(((((((((.(((...(((((...))))).))).))))))))) ( -30.10) >DroAna_CAF1 17584 107 - 1 AACGAGGAGAAAAAAAAAAAAGAGAA-CACCUCGAGCCUUGAGGCGAGUUACCCGCCCAGCAAAAAUUGACAUUGUUCAUAGUCAUAACAUGACGGAAAAAUGUCAAU ..(((((...................-..))))).((..((.((((.......))))))))....(((((((((.(((...(((((...))))).))).))))))))) ( -29.20) >consensus AAA__GGAGAGGGACCAGGAAAA__AAACCUUUGGGCCUUCAAG________GCAUCCAGCUA_AAUUGACAUUGUUCAUAGUCAUAACAUGACGGAAAAAUGUCAAU .....((.(((((...............)))))...))...........................(((((((((.(((...(((((...))))).))).))))))))) (-17.90 = -18.14 + 0.24)

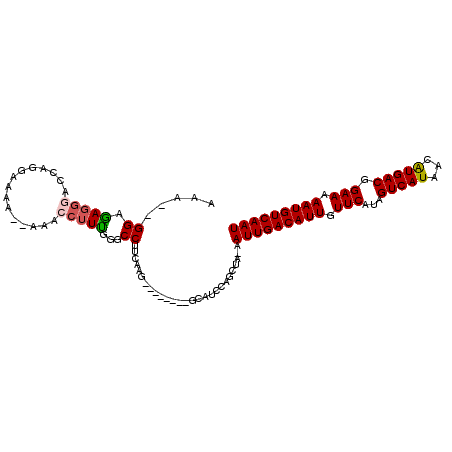
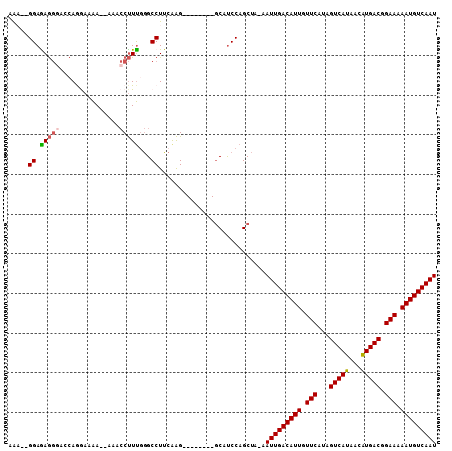
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:46 2006