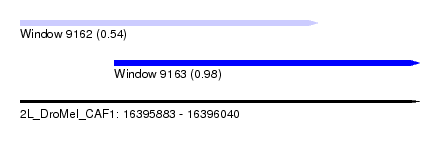
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,395,883 – 16,396,040 |
| Length | 157 |
| Max. P | 0.983973 |

| Location | 16,395,883 – 16,396,000 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.39 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -22.65 |
| Energy contribution | -22.93 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
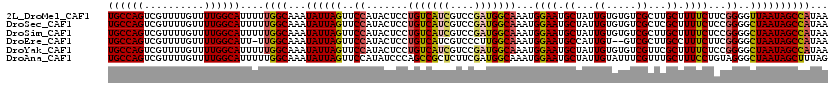
>2L_DroMel_CAF1 16395883 117 + 22407834 ---GUUGUUCCUAAACUGUUUUGUUUUAUUGUGAUUUUUAUGCCAGUCGUUUUGUUUUGGCAUUUUUGGCAAAUAUUAGUUCCAUACUCCUGUCAUCGUCCGAUGGCAAAUGGAAUGCUA ---.........((((......))))..((((.(.....(((((((..........)))))))...).))))......(((((((.....(((((((....))))))).))))))).... ( -25.20) >DroSec_CAF1 20294 117 + 1 ---GUUGUUCCUACACUGUUUUGUUUUAUUGUGAUUUUUAUGCCAGUCGUUUUGUUUUGGCAUUUUUGGCAAAUAUUAGUUCCAUACUCCUGUCAUCGUCCGAUGGCAAAUGGAAUGCUA ---.........................((((.(.....(((((((..........)))))))...).))))......(((((((.....(((((((....))))))).))))))).... ( -24.80) >DroSim_CAF1 18808 117 + 1 ---GUUGUUCCUACACUGUUUUGUUUUAUUGUGAUUUUUAUGCCAGUCGUUUUGUUUUGGCAUUUUUGGCAAAUAUUAGUUCCAUACUCCUGUCAUCGUCCGAUGGCAAAUGGAAUGCUA ---.........................((((.(.....(((((((..........)))))))...).))))......(((((((.....(((((((....))))))).))))))).... ( -24.80) >DroEre_CAF1 16796 116 + 1 ---GUUGUUCCUGCUCUGUUUUGUUUUAUUGUGAUUUUUAUGCCAGUCGUUUUGUUUUGGCAUU-UUGGCAAAUAUUAGUUCCAUACUCCUGUCAUCGUCCCUUGGCAAAUGGAAUGCCA ---.........((..............((((.(.....(((((((..........))))))).-.).))))......(((((((.....(((((........))))).))))))))).. ( -20.80) >DroYak_CAF1 20850 117 + 1 ---GUUGUUCCUGCCCUGUUUUGUUUUAUUGUGAUUUUUAUGCCAGUCGUUUUGUUUUGGCAUUUUUGGCAAAUAUUAGUUCCAUACUCCUGUCAUCGUCCGAUGGCAAAUGGAAUGCUA ---.........((..............((((.(.....(((((((..........)))))))...).))))......(((((((.....(((((((....))))))).))))))))).. ( -25.60) >DroAna_CAF1 15901 120 + 1 GCUGUUGUUCUUGGAUUGUUUUGUUUUAUUGUGAUUUUUAUGCCAGUCGUUUUGUUUUGGCAUUUUUGGCAAAUAUUAGUUCCAUAUCCCAGCCGCUCUUCGAUGGCAAAUGGAAUGCUA ((((..((...((((......((((....(((.(.....(((((((..........)))))))...).))))))).....)))).))..)))).((..(((.((.....)).))).)).. ( -22.40) >consensus ___GUUGUUCCUACACUGUUUUGUUUUAUUGUGAUUUUUAUGCCAGUCGUUUUGUUUUGGCAUUUUUGGCAAAUAUUAGUUCCAUACUCCUGUCAUCGUCCGAUGGCAAAUGGAAUGCUA ............................((((.(.....(((((((..........)))))))...).))))......(((((((.....(((((((....))))))).))))))).... (-22.65 = -22.93 + 0.28)


| Location | 16,395,920 – 16,396,040 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.61 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -30.55 |
| Energy contribution | -30.75 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
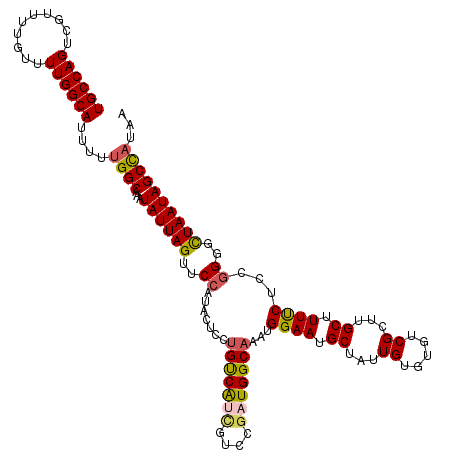
>2L_DroMel_CAF1 16395920 120 + 22407834 UGCCAGUCGUUUUGUUUUGGCAUUUUUGGCAAAUAUUAGUUCCAUACUCCUGUCAUCGUCCGAUGGCAAAUGGAAUGCUAUUGUGUGUCGCUUGCUUUUCUUCGGGGUUAAUAGCCAUAA ((((((..........))))))....((((...((((((..((.......(((((((....)))))))...((((.((....(((...)))..)).))))...))..))))))))))... ( -32.90) >DroSec_CAF1 20331 120 + 1 UGCCAGUCGUUUUGUUUUGGCAUUUUUGGCAAAUAUUAGUUCCAUACUCCUGUCAUCGUCCGAUGGCAAAUGGAAUGCUAUUGUGUGUCGCUCGCUUUUCUCCGGGGCUAAUAGCCAUAA ((((((..........))))))....((((...((((((((((.......(((((((....)))))))...(((..((....))(((.....))).....)))))))))))))))))... ( -35.50) >DroSim_CAF1 18845 120 + 1 UGCCAGUCGUUUUGUUUUGGCAUUUUUGGCAAAUAUUAGUUCCAUACUCCUGUCAUCGUCCGAUGGCAAAUGGAAUGCUAUUGUGUGUCGCUUGCUUUUCUCCGGGGCUAAUAGCCAUAA ((((((..........))))))....((((...((((((((((.......(((((((....)))))))...((((.((....(((...)))..)).))))...))))))))))))))... ( -35.80) >DroEre_CAF1 16833 117 + 1 UGCCAGUCGUUUUGUUUUGGCAUU-UUGGCAAAUAUUAGUUCCAUACUCCUGUCAUCGUCCCUUGGCAAAUGGAAUGCCAUUGU--GUCGCUUGCCUUUCUUCGGGGCUAAUAGCCAUAA ((((((..........))))))..-.((((...((((((((((.......(((((........)))))(((((....)))))..--.................))))))))))))))... ( -31.20) >DroYak_CAF1 20887 120 + 1 UGCCAGUCGUUUUGUUUUGGCAUUUUUGGCAAAUAUUAGUUCCAUACUCCUGUCAUCGUCCGAUGGCAAAUGGAAUGCUAUUGUGUGUCGUUCGCUUUUCUCCGGGGCUAAUAGCCAUAA ((((((..........))))))....((((...((((((((((.......(((((((....)))))))....((((((........).)))))..........))))))))))))))... ( -35.80) >DroAna_CAF1 15941 120 + 1 UGCCAGUCGUUUUGUUUUGGCAUUUUUGGCAAAUAUUAGUUCCAUAUCCCAGCCGCUCUUCGAUGGCAAAUGGAAUGCUAUUGUAUUUCGUUUGCUUUCCUGUAGGGCUAAUAGCUUUAG ((((((..........)))))).....(((.....................)))(((((..((.((((((((((((((....)))))))))))))).))....)))))............ ( -34.00) >consensus UGCCAGUCGUUUUGUUUUGGCAUUUUUGGCAAAUAUUAGUUCCAUACUCCUGUCAUCGUCCGAUGGCAAAUGGAAUGCUAUUGUGUGUCGCUUGCUUUUCUCCGGGGCUAAUAGCCAUAA ((((((..........))))))....((((...((((((..((.......(((((((....)))))))...((((.((...((.....))...)).))))...))..))))))))))... (-30.55 = -30.75 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:40 2006