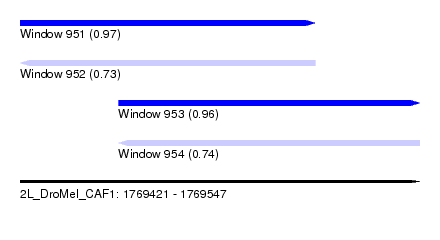
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,769,421 – 1,769,547 |
| Length | 126 |
| Max. P | 0.965760 |

| Location | 1,769,421 – 1,769,514 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.65 |
| Mean single sequence MFE | -27.69 |
| Consensus MFE | -24.60 |
| Energy contribution | -24.10 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1769421 93 + 22407834 AUCUUAGGAAUCAU---------UGUUUCAUCGAAAUUUGCAUAAUUCGCUGCGAAUUUUGUGGUCGGAAUUUAACGAGCCACACAAUUUGUCAGCGGACAA------------------ ......(((((...---------.)))))................(((((((((((((.((((((((........))).))))).)))))).)))))))...------------------ ( -27.70) >DroPse_CAF1 29773 108 + 1 AUCUUAAAGAUC------------GUUUCAUUCAAAUUUACAUAAUUCGUUGCAAGUUUUGGGGCCAACAUUUAACGAGCCCCACAAUUUGUCAGCGGACAGCGGACAAGGCAUUCCACA ............------------.....................(((((((((((((.(((((((..........).)))))).)))))).)))))))..(.(((........))).). ( -27.00) >DroSim_CAF1 29269 93 + 1 AUCUUAGGAAUCAU---------UGUUUCAUUGAAAUUUGCAUAAUUCGCUGCGAAUUUUGUGGUCGGAAUUUAACGAGCCACACAAUUUGUCAGCGGACAA------------------ ......(((((...---------.)))))................(((((((((((((.((((((((........))).))))).)))))).)))))))...------------------ ( -27.70) >DroEre_CAF1 26241 93 + 1 GUCUUAAGAAUCAU---------UGUUUCAUUGAAAUUUGCAUAAUUCGCUGCGAAUUUUGUGGUCGGAAUUUAACGAGCCACACAAUUUGUCAGCGGACAA------------------ ((((...((((...---------(((.............)))..))))((((((((((.((((((((........))).))))).)))))).))))))))..------------------ ( -28.42) >DroYak_CAF1 35401 93 + 1 GACUUGAGAAUCAU---------UGUUUCAUUGAAAUUUGCAUAAUUCGCUGCGAAUUUUGUGGUCGCAAUUUAACGAGCCACACAAUUUGUCAGCGGACAA------------------ ....(((((.....---------..)))))...............(((((((((((((.((((((((........))).))))).)))))).)))))))...------------------ ( -26.10) >DroAna_CAF1 25744 102 + 1 AUCUGCGGGAUCGUCGUCCUGUCUGUUUCAUUGAAAUUUGCAUAAUUCGCUGGGAGUUUUGUGGCCGGAAUUUAACGAGCCAAACAAUUUGUCAGCGGACGG------------------ ....(((....)))((((((((..(((((...)))))..)))......((((((((((...((((((........)).))))...))))).)))))))))).------------------ ( -29.20) >consensus AUCUUAAGAAUCAU_________UGUUUCAUUGAAAUUUGCAUAAUUCGCUGCGAAUUUUGUGGUCGGAAUUUAACGAGCCACACAAUUUGUCAGCGGACAA__________________ .............................................(((((((((((((.((((((((........)).)))))).)))))).)))))))..................... (-24.60 = -24.10 + -0.50)

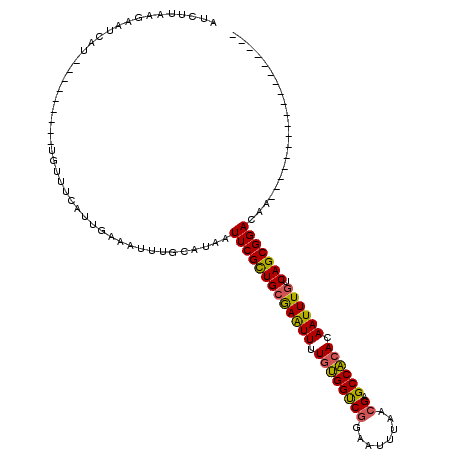
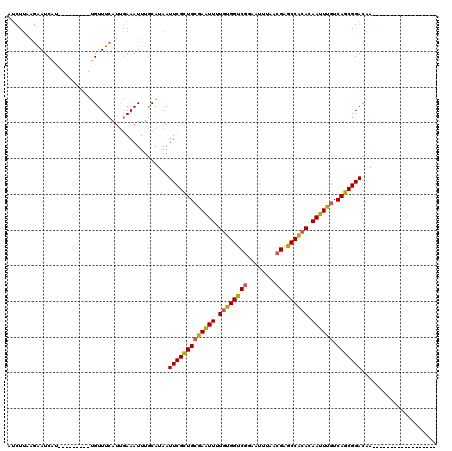
| Location | 1,769,421 – 1,769,514 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.65 |
| Mean single sequence MFE | -22.18 |
| Consensus MFE | -16.72 |
| Energy contribution | -16.75 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1769421 93 - 22407834 ------------------UUGUCCGCUGACAAAUUGUGUGGCUCGUUAAAUUCCGACCACAAAAUUCGCAGCGAAUUAUGCAAAUUUCGAUGAAACA---------AUGAUUCCUAAGAU ------------------......((((.(.((((.(((((.(((........)))))))).)))).)))))(((((((.....((((...))))..---------)))))))....... ( -21.00) >DroPse_CAF1 29773 108 - 1 UGUGGAAUGCCUUGUCCGCUGUCCGCUGACAAAUUGUGGGGCUCGUUAAAUGUUGGCCCCAAAACUUGCAACGAAUUAUGUAAAUUUGAAUGAAAC------------GAUCUUUAAGAU .(((((..((.......))..)))))((.(((.((.((((((.((........)))))))).)).))))).((((((.....))))))........------------............ ( -23.90) >DroSim_CAF1 29269 93 - 1 ------------------UUGUCCGCUGACAAAUUGUGUGGCUCGUUAAAUUCCGACCACAAAAUUCGCAGCGAAUUAUGCAAAUUUCAAUGAAACA---------AUGAUUCCUAAGAU ------------------......((((.(.((((.(((((.(((........)))))))).)))).)))))(((((((.....((((...))))..---------)))))))....... ( -21.00) >DroEre_CAF1 26241 93 - 1 ------------------UUGUCCGCUGACAAAUUGUGUGGCUCGUUAAAUUCCGACCACAAAAUUCGCAGCGAAUUAUGCAAAUUUCAAUGAAACA---------AUGAUUCUUAAGAC ------------------..(((.((((.(.((((.(((((.(((........)))))))).)))).)))))(((((((.....((((...))))..---------)))))))....))) ( -22.40) >DroYak_CAF1 35401 93 - 1 ------------------UUGUCCGCUGACAAAUUGUGUGGCUCGUUAAAUUGCGACCACAAAAUUCGCAGCGAAUUAUGCAAAUUUCAAUGAAACA---------AUGAUUCUCAAGUC ------------------......((((.(.((((.(((((.((((......))))))))).)))).)))))(((((((.....((((...))))..---------)))))))....... ( -23.90) >DroAna_CAF1 25744 102 - 1 ------------------CCGUCCGCUGACAAAUUGUUUGGCUCGUUAAAUUCCGGCCACAAAACUCCCAGCGAAUUAUGCAAAUUUCAAUGAAACAGACAGGACGACGAUCCCGCAGAU ------------------.(((((.(((....((((..((((.((........))))))...........(((.....)))......))))....)))...))))).............. ( -20.90) >consensus __________________UUGUCCGCUGACAAAUUGUGUGGCUCGUUAAAUUCCGACCACAAAAUUCGCAGCGAAUUAUGCAAAUUUCAAUGAAACA_________AUGAUUCCUAAGAU .......................(((((...((((.(((((.(((........)))))))).))))..)))))............................................... (-16.72 = -16.75 + 0.03)
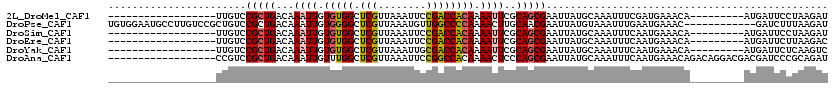

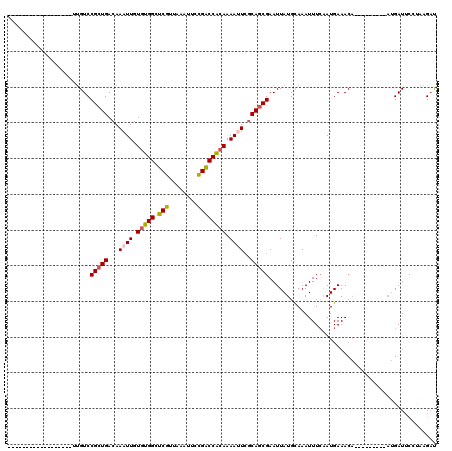
| Location | 1,769,452 – 1,769,547 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.95 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -27.55 |
| Energy contribution | -27.33 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1769452 95 + 22407834 CAUAAUUCGCUGCGAAUUUUGUGGUCGGAAUUUAACGAGCCACACAAUUUGUCAGCGGACAA--------------------CCUCGAAUUAUUCAUGGGCAGCCAAAAAGGCAG .(((((((((((((((((.((((((((........))).))))).)))))).))))((....--------------------))..))))))).........(((.....))).. ( -32.60) >DroSim_CAF1 29300 95 + 1 CAUAAUUCGCUGCGAAUUUUGUGGUCGGAAUUUAACGAGCCACACAAUUUGUCAGCGGACAA--------------------CCUCGAAUUAUUCAUGGGCAGCCAAAAAGGCAG .(((((((((((((((((.((((((((........))).))))).)))))).))))((....--------------------))..))))))).........(((.....))).. ( -32.60) >DroEre_CAF1 26272 95 + 1 CAUAAUUCGCUGCGAAUUUUGUGGUCGGAAUUUAACGAGCCACACAAUUUGUCAGCGGACAA--------------------CCUCGAAUUAUUCAUGGGCAGCCAAAAAGGCAG .(((((((((((((((((.((((((((........))).))))).)))))).))))((....--------------------))..))))))).........(((.....))).. ( -32.60) >DroYak_CAF1 35432 95 + 1 CAUAAUUCGCUGCGAAUUUUGUGGUCGCAAUUUAACGAGCCACACAAUUUGUCAGCGGACAA--------------------CCUCGAAUUAUUCAUGGGCAGCCAAAAAGGCAG .(((((((((((((((((.((((((((........))).))))).)))))).))))((....--------------------))..))))))).........(((.....))).. ( -31.30) >DroAna_CAF1 25784 94 + 1 CAUAAUUCGCUGGGAGUUUUGUGGCCGGAAUUUAACGAGCCAAACAAUUUGUCAGCGGACGG--------------------CC-AGAAUUAUUCAUGGCCACCCCAAGGGGGAG .....(((((((((((((...((((((........)).))))...))))).)))))))).((--------------------((-(((.....)).))))).((((...)))).. ( -36.00) >DroPer_CAF1 28702 109 + 1 CAUAAUUCGUUGCAAGUUUUGGGGCCAACAUUUAACGAGCCCCACAAUUUGUCAGCGGACAGCGGACAAGGCAUUCCACAGUCCUCGAAUUAUUCAUGGGAA------AAGGAGG .(((((((((((((((((.(((((((..........).)))))).)))))).))))((((.(.(((........))).).))))..))))))).........------....... ( -34.90) >consensus CAUAAUUCGCUGCGAAUUUUGUGGUCGGAAUUUAACGAGCCACACAAUUUGUCAGCGGACAA____________________CCUCGAAUUAUUCAUGGGCAGCCAAAAAGGCAG .....(((((((((((((.((((((((........)).)))))).)))))).))))))).......................((..((.....))..))...(((.....))).. (-27.55 = -27.33 + -0.22)

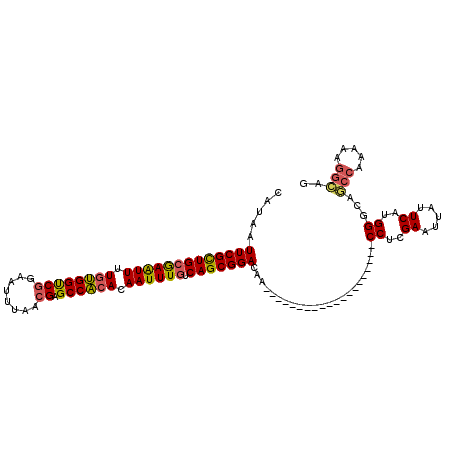
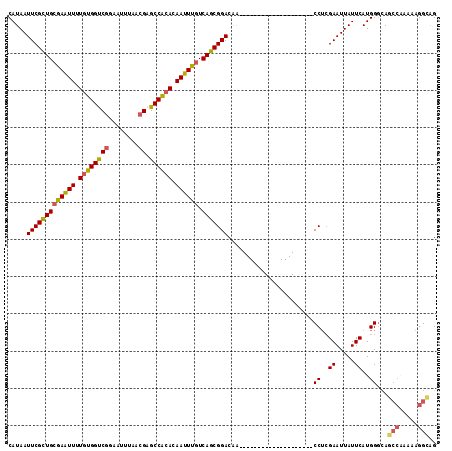
| Location | 1,769,452 – 1,769,547 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.95 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -20.85 |
| Energy contribution | -21.16 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1769452 95 - 22407834 CUGCCUUUUUGGCUGCCCAUGAAUAAUUCGAGG--------------------UUGUCCGCUGACAAAUUGUGUGGCUCGUUAAAUUCCGACCACAAAAUUCGCAGCGAAUUAUG ..(((.....))).........(((((((..((--------------------....))((((.(.((((.(((((.(((........)))))))).)))).)))))))))))). ( -28.60) >DroSim_CAF1 29300 95 - 1 CUGCCUUUUUGGCUGCCCAUGAAUAAUUCGAGG--------------------UUGUCCGCUGACAAAUUGUGUGGCUCGUUAAAUUCCGACCACAAAAUUCGCAGCGAAUUAUG ..(((.....))).........(((((((..((--------------------....))((((.(.((((.(((((.(((........)))))))).)))).)))))))))))). ( -28.60) >DroEre_CAF1 26272 95 - 1 CUGCCUUUUUGGCUGCCCAUGAAUAAUUCGAGG--------------------UUGUCCGCUGACAAAUUGUGUGGCUCGUUAAAUUCCGACCACAAAAUUCGCAGCGAAUUAUG ..(((.....))).........(((((((..((--------------------....))((((.(.((((.(((((.(((........)))))))).)))).)))))))))))). ( -28.60) >DroYak_CAF1 35432 95 - 1 CUGCCUUUUUGGCUGCCCAUGAAUAAUUCGAGG--------------------UUGUCCGCUGACAAAUUGUGUGGCUCGUUAAAUUGCGACCACAAAAUUCGCAGCGAAUUAUG ..(((.....))).........(((((((..((--------------------....))((((.(.((((.(((((.((((......))))))))).)))).)))))))))))). ( -31.10) >DroAna_CAF1 25784 94 - 1 CUCCCCCUUGGGGUGGCCAUGAAUAAUUCU-GG--------------------CCGUCCGCUGACAAAUUGUUUGGCUCGUUAAAUUCCGGCCACAAAACUCCCAGCGAAUUAUG ..((((...)))).(((((.((.....)))-))--------------------))...(((((.....((((..(((.((........)))))))))......)))))....... ( -27.60) >DroPer_CAF1 28702 109 - 1 CCUCCUU------UUCCCAUGAAUAAUUCGAGGACUGUGGAAUGCCUUGUCCGCUGUCCGCUGACAAAUUGUGGGGCUCGUUAAAUGUUGGCCCCAAAACUUGCAACGAAUUAUG .......------.........((((((((.((((.(((((........))))).))))..((.(((.((.((((((.((........)))))))).)).))))).)))))))). ( -35.70) >consensus CUGCCUUUUUGGCUGCCCAUGAAUAAUUCGAGG____________________UUGUCCGCUGACAAAUUGUGUGGCUCGUUAAAUUCCGACCACAAAAUUCGCAGCGAAUUAUG ..(((.....))).........(((((((..((........................))((((...((((.(((((.(((........)))))))).))))..))))))))))). (-20.85 = -21.16 + 0.31)
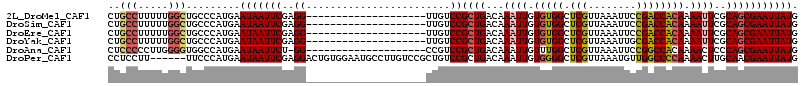
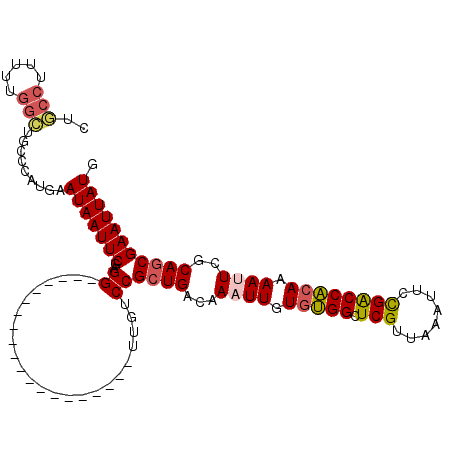
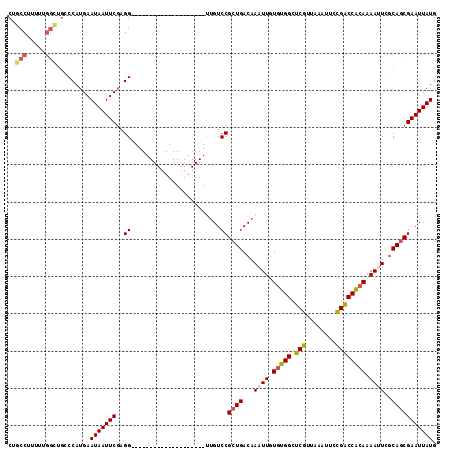
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:09 2006