
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,394,800 – 16,394,971 |
| Length | 171 |
| Max. P | 0.996526 |

| Location | 16,394,800 – 16,394,897 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.74 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -24.20 |
| Energy contribution | -24.45 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16394800 97 + 22407834 UGCAACUGCAUAUUUUGUUUGACAUCAUUGUUGCACAUGCCACGUGC---------------UGCAGUUGCAC------CUGCGCCUGCAAAC--UUUCGCAACAUUGCGGAAAAACGCC ((((((((((..........((((....))))((((.......))))---------------)))))))))).------.(((....)))...--(((((((....)))))))....... ( -30.10) >DroSec_CAF1 19219 96 + 1 GGCAACUGCAUAUUUUGUUAGACAUCAUUGUUGCACAUGCCACGUGC---------------UGCAGUUGCAC------CUGCGCCUGCA-AC--AUUCGCAACAUUGCGGAAAAACGCC .(((((((((..........((((....))))((((.......))))---------------)))))))))..------.(((....)))-..--.((((((....))))))........ ( -29.10) >DroSim_CAF1 17757 96 + 1 UGCAACUGCAUAUUUUGUUUGACAUCAUUGUUGCACAUGCCACGUGC---------------UGCAGUUGCAC------CUGCGCCUGCA-AC--AUUCGCAACAUUGCGGAAAAACGCC ((((((((((..........((((....))))((((.......))))---------------)))))))))).------.(((....)))-..--.((((((....))))))........ ( -29.40) >DroEre_CAF1 15746 95 + 1 UGCAACUGCACAUUUUGCUUGACAUCAUUGUUGCACUUGCCACGUGC---------------UGCAGUUGCAC------CUGCGCCUGCA-AC--AUUCGCAACAGUGCGGAA-AACGCC ((((((((((..........((((....))))((((.......))))---------------)))))))))).------((((((.(((.-..--....)))...))))))..-...... ( -31.70) >DroYak_CAF1 19748 102 + 1 UGCAACUGCGUAUUUUGUUUGACAUCAUUGUUGCACAUGCCACGUGC---------------UGCAGUUGCACCUGCACCUGCGCCUGCA-AC--AUUCGCAACAUUGCGGAAAAACGCC (((((((((...........((((....))))((((.......))))---------------.)))))))))...((...(((....)))-..--.((((((....)))))).....)). ( -30.40) >DroAna_CAF1 15012 110 + 1 UCCAACUGCAUAUUUUGUUUGACAUCAUUGUUGCACAUGCCACGUGCUGGUGGUGUUGCUGCUGCAGUUACUG------UUGCACCUGCA-ACACAUUCGUU--GG-GGGGGAAAACGCC ((((((((((..........((((....))))(((.(((((((......))))))))))...)))))))....------.....(((.((-((......)))--).-))))))....... ( -34.20) >consensus UGCAACUGCAUAUUUUGUUUGACAUCAUUGUUGCACAUGCCACGUGC_______________UGCAGUUGCAC______CUGCGCCUGCA_AC__AUUCGCAACAUUGCGGAAAAACGCC .(((((((((..........((((....))))((((.......))))...............))))))))).........(((....)))......((((((....))))))........ (-24.20 = -24.45 + 0.25)



| Location | 16,394,800 – 16,394,897 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.74 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -24.03 |
| Energy contribution | -24.75 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16394800 97 - 22407834 GGCGUUUUUCCGCAAUGUUGCGAAA--GUUUGCAGGCGCAG------GUGCAACUGCA---------------GCACGUGGCAUGUGCAACAAUGAUGUCAAACAAAAUAUGCAGUUGCA .((((((....((((....((....--))))))))))))..------.((((((((((---------------((((((...)))))).....((....)).........)))))))))) ( -32.40) >DroSec_CAF1 19219 96 - 1 GGCGUUUUUCCGCAAUGUUGCGAAU--GU-UGCAGGCGCAG------GUGCAACUGCA---------------GCACGUGGCAUGUGCAACAAUGAUGUCUAACAAAAUAUGCAGUUGCC ((((((.....(((((((..((..(--((-(((((..((..------..))..)))))---------------)))))..)))).)))......))))))...........((....)). ( -32.30) >DroSim_CAF1 17757 96 - 1 GGCGUUUUUCCGCAAUGUUGCGAAU--GU-UGCAGGCGCAG------GUGCAACUGCA---------------GCACGUGGCAUGUGCAACAAUGAUGUCAAACAAAAUAUGCAGUUGCA ((((((.....(((((((..((..(--((-(((((..((..------..))..)))))---------------)))))..)))).)))......))))))..........(((....))) ( -33.90) >DroEre_CAF1 15746 95 - 1 GGCGUU-UUCCGCACUGUUGCGAAU--GU-UGCAGGCGCAG------GUGCAACUGCA---------------GCACGUGGCAAGUGCAACAAUGAUGUCAAGCAAAAUGUGCAGUUGCA ((((((-....(((((((..((..(--((-(((((..((..------..))..)))))---------------)))))..)).)))))......))))))..((((.........)))). ( -36.50) >DroYak_CAF1 19748 102 - 1 GGCGUUUUUCCGCAAUGUUGCGAAU--GU-UGCAGGCGCAGGUGCAGGUGCAACUGCA---------------GCACGUGGCAUGUGCAACAAUGAUGUCAAACAAAAUACGCAGUUGCA .((((.(((...((.(((((((.((--((-..(..((((((.(((....))).)))).---------------))..)..)))).))))))).)).((.....))))).))))....... ( -35.60) >DroAna_CAF1 15012 110 - 1 GGCGUUUUCCCCC-CC--AACGAAUGUGU-UGCAGGUGCAA------CAGUAACUGCAGCAGCAACACCACCAGCACGUGGCAUGUGCAACAAUGAUGUCAAACAAAAUAUGCAGUUGGA .............-((--(((....((((-(((.(.((((.------.......)))).).)))))))(((......)))((((((.......((....))......)))))).))))). ( -30.02) >consensus GGCGUUUUUCCGCAAUGUUGCGAAU__GU_UGCAGGCGCAG______GUGCAACUGCA_______________GCACGUGGCAUGUGCAACAAUGAUGUCAAACAAAAUAUGCAGUUGCA .((((((...((((....)))).....(....)))))))..........((((((((................((((((...))))))........((.....))......)))))))). (-24.03 = -24.75 + 0.72)

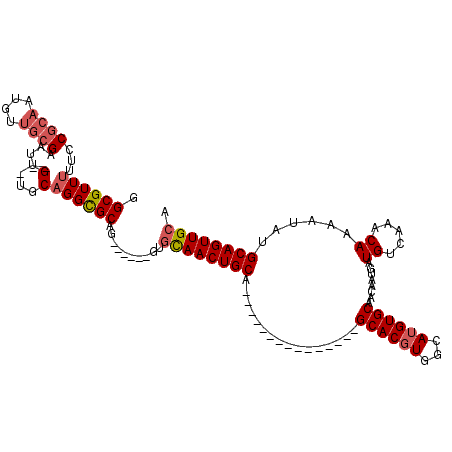

| Location | 16,394,840 – 16,394,931 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.53 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -21.90 |
| Energy contribution | -22.15 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16394840 91 + 22407834 CACGUGC---------------UGCAGUUGCAC------CUGCGCCUGCAAAC--UUUCGCAACAUUGCGGAAAAACGCCCCAAAAAGGCGGCAACAUGCAACA----GUGG--GGAAUC ..(((..---------------(((((.(((..------..))).)))))...--(((((((....)))))))..)))(((((.....(((......)))....----.)))--)).... ( -30.70) >DroSec_CAF1 19259 90 + 1 CACGUGC---------------UGCAGUUGCAC------CUGCGCCUGCA-AC--AUUCGCAACAUUGCGGAAAAACGCCCCAAAAAGGCGGCAACAUGCAACA----GUGG--GGAAUC ..((((.---------------(((((.(((..------..))).)))))-.)--.((((((....))))))...)))(((((.....(((......)))....----.)))--)).... ( -30.20) >DroSim_CAF1 17797 90 + 1 CACGUGC---------------UGCAGUUGCAC------CUGCGCCUGCA-AC--AUUCGCAACAUUGCGGAAAAACGCCCCAAAAAGGCGGCAACAUGCAACA----GUGG--GGAAUC ..((((.---------------(((((.(((..------..))).)))))-.)--.((((((....))))))...)))(((((.....(((......)))....----.)))--)).... ( -30.20) >DroEre_CAF1 15786 89 + 1 CACGUGC---------------UGCAGUUGCAC------CUGCGCCUGCA-AC--AUUCGCAACAGUGCGGAA-AACGCCCCAAAAAGGCGGCAACAUGCAACA----GUGG--GCAGUC (((.((.---------------(((((((((..------((((((.(((.-..--....)))...))))))..-..((((.......))))))))).)))).))----))).--...... ( -32.40) >DroYak_CAF1 19788 95 + 1 CACGUGC---------------UGCAGUUGCACCUGCACCUGCGCCUGCA-AC--AUUCGCAACAUUGCGGAAAAACGCCCCAAAAAGGCGGCAACAUGCAACA----GUAG--G-AGUC ..(.(((---------------(((((.(((....))).))))...((((-..--.((((((....)))))).....(((((.....)).)))....))))..)----))).--)-.... ( -31.40) >DroAna_CAF1 15052 110 + 1 CACGUGCUGGUGGUGUUGCUGCUGCAGUUACUG------UUGCACCUGCA-ACACAUUCGUU--GG-GGGGGAAAACGCCCCAAAAAGGCGGCAACAUGCAACGCGCCGUCGAAGGCCUU (((......)))(((((((((((((......((------((((....)))-)))......((--((-((.(.....).))))))....))))))....)))))))(((......)))... ( -44.90) >consensus CACGUGC_______________UGCAGUUGCAC______CUGCGCCUGCA_AC__AUUCGCAACAUUGCGGAAAAACGCCCCAAAAAGGCGGCAACAUGCAACA____GUGG__GGAAUC ..((((................(((((.((((........)))).)))))......((((((....)))))).....(((((.....)).)))..))))..................... (-21.90 = -22.15 + 0.25)
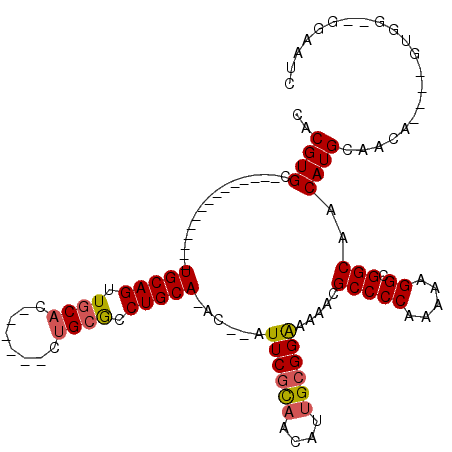
| Location | 16,394,840 – 16,394,931 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.53 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -22.54 |
| Energy contribution | -23.07 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16394840 91 - 22407834 GAUUCC--CCAC----UGUUGCAUGUUGCCGCCUUUUUGGGGCGUUUUUCCGCAAUGUUGCGAAA--GUUUGCAGGCGCAG------GUGCAACUGCA---------------GCACGUG ......--.(((----(((((((.((((((((((.....))))).....((((..(((.((....--))..)))...)).)------).)))))))))---------------))).))) ( -34.00) >DroSec_CAF1 19259 90 - 1 GAUUCC--CCAC----UGUUGCAUGUUGCCGCCUUUUUGGGGCGUUUUUCCGCAAUGUUGCGAAU--GU-UGCAGGCGCAG------GUGCAACUGCA---------------GCACGUG ......--.(((----(((((((.(((((.((((.((((.((((((....((((....)))))))--))-).))))...))------)))))))))))---------------))).))) ( -33.40) >DroSim_CAF1 17797 90 - 1 GAUUCC--CCAC----UGUUGCAUGUUGCCGCCUUUUUGGGGCGUUUUUCCGCAAUGUUGCGAAU--GU-UGCAGGCGCAG------GUGCAACUGCA---------------GCACGUG ......--.(((----(((((((.(((((.((((.((((.((((((....((((....)))))))--))-).))))...))------)))))))))))---------------))).))) ( -33.40) >DroEre_CAF1 15786 89 - 1 GACUGC--CCAC----UGUUGCAUGUUGCCGCCUUUUUGGGGCGUU-UUCCGCACUGUUGCGAAU--GU-UGCAGGCGCAG------GUGCAACUGCA---------------GCACGUG ......--.(((----(((((((.((((((((((.....)))))..-...(((.((((.((....--))-.)))))))...------..)))))))))---------------))).))) ( -34.90) >DroYak_CAF1 19788 95 - 1 GACU-C--CUAC----UGUUGCAUGUUGCCGCCUUUUUGGGGCGUUUUUCCGCAAUGUUGCGAAU--GU-UGCAGGCGCAGGUGCAGGUGCAACUGCA---------------GCACGUG ....-.--.(((----(((((((.((((((((((.....))))).....(((((...(((((..(--(.-..))..))))).))).)).)))))))))---------------))).))) ( -34.60) >DroAna_CAF1 15052 110 - 1 AAGGCCUUCGACGGCGCGUUGCAUGUUGCCGCCUUUUUGGGGCGUUUUCCCCC-CC--AACGAAUGUGU-UGCAGGUGCAA------CAGUAACUGCAGCAGCAACACCACCAGCACGUG ...(((......)))(.((((..((((((.......((((((.(......)))-))--))......(((-(((((.(((..------..))).))))))))))))))....)))).)... ( -37.30) >consensus GAUUCC__CCAC____UGUUGCAUGUUGCCGCCUUUUUGGGGCGUUUUUCCGCAAUGUUGCGAAU__GU_UGCAGGCGCAG______GUGCAACUGCA_______________GCACGUG .....................(((((.(((((((.....)))))......((((....))))........(((((..(((........)))..)))))...............))))))) (-22.54 = -23.07 + 0.53)



| Location | 16,394,859 – 16,394,971 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.53 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -33.00 |
| Energy contribution | -33.08 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16394859 112 + 22407834 UGCGCCUGCAAAC--UUUCGCAACAUUGCGGAAAAACGCCCCAAAAAGGCGGCAACAUGCAACA----GUGG--GGAAUCGCAAAAAAUGCAGCCAGCUGCAUGUGCGUGCAUAAAUUUA (((((........--(((((((....))))))).....(((((.....(((......)))....----.)))--))....(((....((((((....)))))).))))))))........ ( -37.50) >DroSec_CAF1 19278 111 + 1 UGCGCCUGCA-AC--AUUCGCAACAUUGCGGAAAAACGCCCCAAAAAGGCGGCAACAUGCAACA----GUGG--GGAAUCGCAAAAAAUGCAGCCAGCUGCAUGUGCGUGCAUAAAUUUA (((((.....-..--.((((((....))))))......(((((.....(((......)))....----.)))--))....(((....((((((....)))))).))))))))........ ( -36.80) >DroSim_CAF1 17816 111 + 1 UGCGCCUGCA-AC--AUUCGCAACAUUGCGGAAAAACGCCCCAAAAAGGCGGCAACAUGCAACA----GUGG--GGAAUCGCAAAAAAUGCAGCCAGCUGCAUGUGCGUGCAUAAAUUUA (((((.....-..--.((((((....))))))......(((((.....(((......)))....----.)))--))....(((....((((((....)))))).))))))))........ ( -36.80) >DroEre_CAF1 15805 110 + 1 UGCGCCUGCA-AC--AUUCGCAACAGUGCGGAA-AACGCCCCAAAAAGGCGGCAACAUGCAACA----GUGG--GCAGUCGCAAAAAAUGCAGCCAGCUGCAUGUGCGUGCAUAAAUUUA ((((((..(.-..--.((((((....)))))).-...(((((.....)).)))...........----)..)--))...((((....((((((....)))))).)))).)))........ ( -40.20) >DroYak_CAF1 19813 110 + 1 UGCGCCUGCA-AC--AUUCGCAACAUUGCGGAAAAACGCCCCAAAAAGGCGGCAACAUGCAACA----GUAG--G-AGUCGCAAAAAAUGCAGCCAGCUGCAUGUGCGUGCAUAAAUUUA (((.(((((.-..--.((((((....)))))).....(((((.....)).)))...........----))))--)-...((((....((((((....)))))).)))).)))........ ( -37.30) >DroAna_CAF1 15086 116 + 1 UGCACCUGCA-ACACAUUCGUU--GG-GGGGGAAAACGCCCCAAAAAGGCGGCAACAUGCAACGCGCCGUCGAAGGCCUUGCAAAAAAUGCAACCAGCUGCAUGUGCGUGCAUAAAUUUC (((((.(((.-.(.......((--((-((.(.....).))))))....)..)))(((((((..(((((......))).(((((.....)))))...)))))))))..)))))........ ( -40.60) >consensus UGCGCCUGCA_AC__AUUCGCAACAUUGCGGAAAAACGCCCCAAAAAGGCGGCAACAUGCAACA____GUGG__GGAAUCGCAAAAAAUGCAGCCAGCUGCAUGUGCGUGCAUAAAUUUA (((((.((((......((((((....)))))).....(((((.....)).)))....))))...................(((....((((((....)))))).))))))))........ (-33.00 = -33.08 + 0.09)

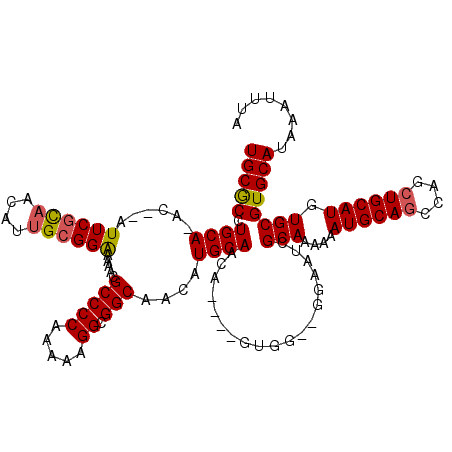
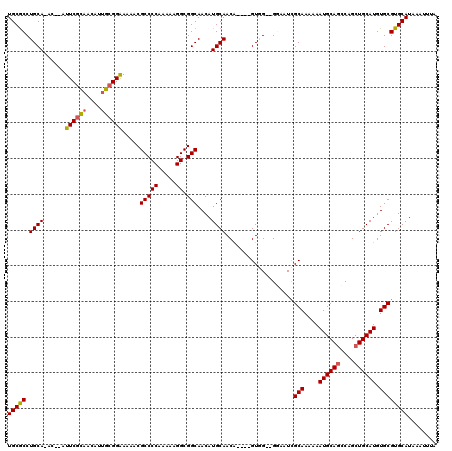
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:36 2006