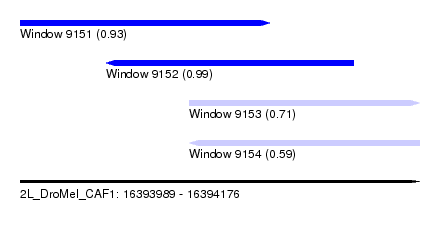
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,393,989 – 16,394,176 |
| Length | 187 |
| Max. P | 0.994877 |

| Location | 16,393,989 – 16,394,106 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 98.46 |
| Mean single sequence MFE | -18.93 |
| Consensus MFE | -17.19 |
| Energy contribution | -17.03 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16393989 117 + 22407834 GCAAAAUGUAUUUUCCCAUUGCAUUUUCGAUUAGUUUUUCUGAUUUGACCAUACAUAUUUGCACAAACAAUCAACAAAAUAAUUAAAUGCAUUCAAGCAUUUUUAAUUGUUAUUUGC (((((((((((..((.............((((((.....)))))).))..)))))).))))).....(((..(((((.......((((((......))))))....)))))..))). ( -17.94) >DroSec_CAF1 18542 117 + 1 GCAGAAUGUAUUUUCCCAUUGCAUUUUCGAUUAGUUUUUCUGAUUUGACCAUACAUAUUUGCACAAACAAUCAACAAAAUAAUUAAAUGCAUUCAAGCAUUUUUAAUUGUUAUUUGC (((((((((((..((.............((((((.....)))))).))..)))))).))))).....(((..(((((.......((((((......))))))....)))))..))). ( -18.04) >DroSim_CAF1 17048 117 + 1 GCAAAAUGUAUUUUCCCAUUGCAUUUUCGAUUAGUUUUUCUGAUUUGACCAUACAUAUUUGCACAAACAAUCAACAAAAUAAUUAAAUGCAUUCAAGCAUUUUUAAUUGUUAUUUGC (((((((((((..((.............((((((.....)))))).))..)))))).))))).....(((..(((((.......((((((......))))))....)))))..))). ( -17.94) >DroEre_CAF1 15197 117 + 1 GCAAAAUGUAUUUUCCCACUGCAUUUUCGAUUAGUUUUUCUGAUUUGACCAUACAUAUUUGCACAAACAAUCAACAAAAUAAUUAAAUGCAUUCAAGCAUUUUUAAUUGUUGUUUGC (((((((((((..((.............((((((.....)))))).))..)))))).))))).....(((.((((((.......((((((......))))))....)))))).))). ( -20.74) >DroYak_CAF1 18999 117 + 1 GCAAAAUGUAUUUCCCCAUUGCAUUUUCGAUUAGUUUUUCUGAUUUGACCAUACAUAUUUGCACAAACAAUCAACAAAAUAAUUAAAUGCAUUCAAGCAUUUUUAAUUGUUGUUUGC ((((((((((.........)))))..((((((((.....)))).)))).........))))).....(((.((((((.......((((((......))))))....)))))).))). ( -20.00) >consensus GCAAAAUGUAUUUUCCCAUUGCAUUUUCGAUUAGUUUUUCUGAUUUGACCAUACAUAUUUGCACAAACAAUCAACAAAAUAAUUAAAUGCAUUCAAGCAUUUUUAAUUGUUAUUUGC (((((((((((...............((((((((.....)))).))))..)))))).))))).....(((..(((((.......((((((......))))))....)))))..))). (-17.19 = -17.03 + -0.16)

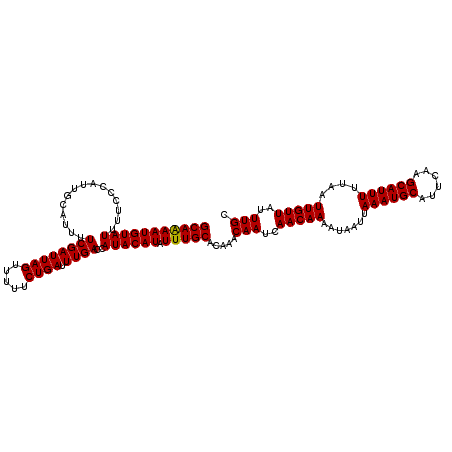
| Location | 16,394,029 – 16,394,145 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 97.16 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -24.04 |
| Energy contribution | -23.64 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16394029 116 - 22407834 UCGCCGCCA-AUCGACGAACGCCUAAGCAUCGAAAUUAGCGCAAAUAACAAUUAAAAAUGCUUGAAUGCAUUUAAUUAUUUUGUUGAUUGUUUGUGCAAAUAUGUAUGGUCAAAUCA ..(((..((-(((((((((.((((((.........)))).)).............((((((......))))))......)))))))))))...(((((....))))))))....... ( -25.80) >DroSec_CAF1 18582 116 - 1 UAGCCGCCA-AUCGACGAACGCCUAAGCACCGAAAUUAGCGCAAAUAACAAUUAAAAAUGCUUGAAUGCAUUUAAUUAUUUUGUUGAUUGUUUGUGCAAAUAUGUAUGGUCAAAUCA ..(((..((-(((((((((.((((((.........)))).)).............((((((......))))))......)))))))))))...(((((....))))))))....... ( -25.80) >DroSim_CAF1 17088 116 - 1 UCGCCGCCA-AUCGACGAACGCCUAAGCACCAAAAUUAGCGCAAAUAACAAUUAAAAAUGCUUGAAUGCAUUUAAUUAUUUUGUUGAUUGUUUGUGCAAAUAUGUAUGGUCAAAUCA ..(((..((-(((((((((.((((((.........)))).)).............((((((......))))))......)))))))))))...(((((....))))))))....... ( -25.80) >DroEre_CAF1 15237 117 - 1 UGGCCGCCAAAUCGACGAACGCCUAAGCAACGAAAUUAGCGCAAACAACAAUUAAAAAUGCUUGAAUGCAUUUAAUUAUUUUGUUGAUUGUUUGUGCAAAUAUGUAUGGUCAAAUCA ((((((..............((....))..........(((((((((((((((((((((((......))))))..)))....)))).)))))))))).........))))))..... ( -27.50) >DroYak_CAF1 19039 116 - 1 UAGCCGCCA-AUCGACGGACGCCUAAGCACCGAAAUUAGCGCAAACAACAAUUAAAAAUGCUUGAAUGCAUUUAAUUAUUUUGUUGAUUGUUUGUGCAAAUAUGUAUGGUCAAAUCA .....((((-.....(((..((....)).)))......(((((((((((((((((((((((......))))))..)))....)))).)))))))))).........))))....... ( -27.60) >consensus UAGCCGCCA_AUCGACGAACGCCUAAGCACCGAAAUUAGCGCAAAUAACAAUUAAAAAUGCUUGAAUGCAUUUAAUUAUUUUGUUGAUUGUUUGUGCAAAUAUGUAUGGUCAAAUCA .............(((.(((((....))..........(((((((((((((((((((((((......))))))..)))....)))).))))))))))......)).).)))...... (-24.04 = -23.64 + -0.40)

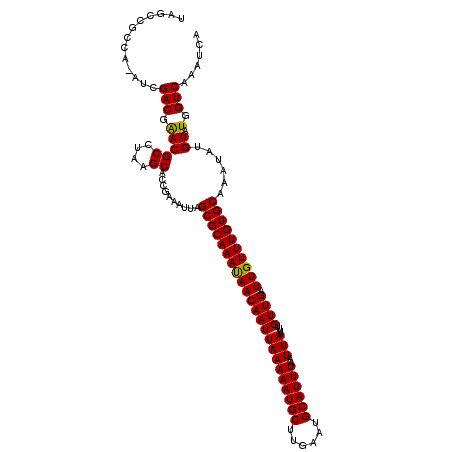
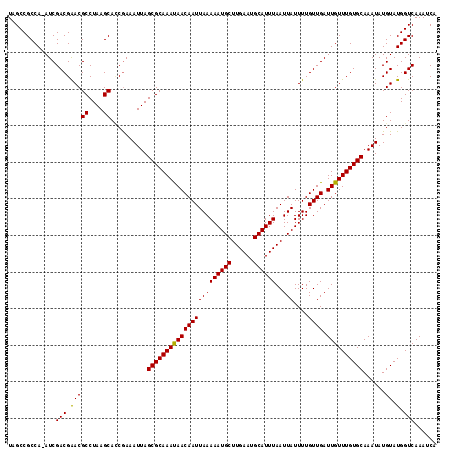
| Location | 16,394,068 – 16,394,176 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 92.68 |
| Mean single sequence MFE | -34.26 |
| Consensus MFE | -26.74 |
| Energy contribution | -26.90 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16394068 108 + 22407834 UAAUUAAAUGCAUUCAAGCAUUUUUAAUUGUUAUUUGCGCUAAUUUCGAUGCUUAGGCGUUCGUCGAU-UGGCGGCGAGU-------GGGCGUGGU--CGCCAGUAAACAGCCUGCAU (((((((((((......))))..))))))).....((((((((((.((((((....))).)))..)))-))))(((..((-------.((((....--)))).....)).))).))). ( -32.60) >DroSec_CAF1 18621 108 + 1 UAAUUAAAUGCAUUCAAGCAUUUUUAAUUGUUAUUUGCGCUAAUUUCGGUGCUUAGGCGUUCGUCGAU-UGGCGGCUAGU-------GGGCGUGGU--CGCCAGUAAAUAGCCUGCAU (((((((((((......))))..)))))))......(((((......))))).(((((........((-((((((((((.-------...).))))--))))))).....)))))... ( -33.02) >DroSim_CAF1 17127 108 + 1 UAAUUAAAUGCAUUCAAGCAUUUUUAAUUGUUAUUUGCGCUAAUUUUGGUGCUUAGGCGUUCGUCGAU-UGGCGGCGAGU-------GGGCGUGGU--CGCCAGUAAACAGCCUGCAU (((((((((((......))))..)))))))......((((((....)))))).(((((....((..((-(((((((.((.-------...).).))--)))))))..)).)))))... ( -33.20) >DroEre_CAF1 15276 118 + 1 UAAUUAAAUGCAUUCAAGCAUUUUUAAUUGUUGUUUGCGCUAAUUUCGUUGCUUAGGCGUUCGUCGAUUUGGCGGCCAGUGGGCAGUGGGCGUGGUCCCGCCCGUAAAUAGCCUGCAU .((((((((((......))))..))))))((((((((((((((..(((.(((....))).....))).))))).(((....)))...(((((......))))))))))))))...... ( -36.10) >DroYak_CAF1 19078 108 + 1 UAAUUAAAUGCAUUCAAGCAUUUUUAAUUGUUGUUUGCGCUAAUUUCGGUGCUUAGGCGUCCGUCGAU-UGGCGGCUAGU-------GGGCGUGGU--CGCCGGUAAACAGCCUGCAU .((((((((((......))))..))))))((((((((((((((((.((((((....))).)))..)))-)))).......-------.((((....--)))).)))))))))...... ( -36.40) >consensus UAAUUAAAUGCAUUCAAGCAUUUUUAAUUGUUAUUUGCGCUAAUUUCGGUGCUUAGGCGUUCGUCGAU_UGGCGGCGAGU_______GGGCGUGGU__CGCCAGUAAACAGCCUGCAU .((((((((((......))))..))))))(((((((((((((...(((((((....)))....))))..))))...............((((......)))).)))))))))...... (-26.74 = -26.90 + 0.16)

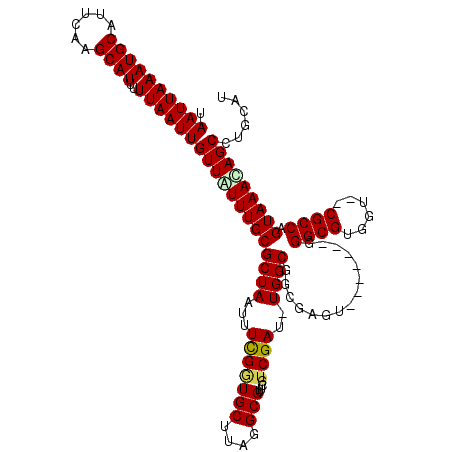
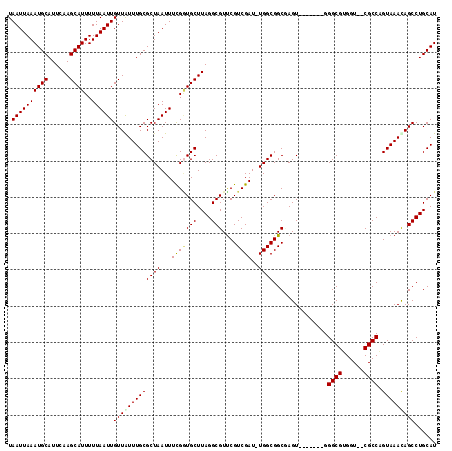
| Location | 16,394,068 – 16,394,176 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 92.68 |
| Mean single sequence MFE | -25.12 |
| Consensus MFE | -19.88 |
| Energy contribution | -19.88 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16394068 108 - 22407834 AUGCAGGCUGUUUACUGGCG--ACCACGCCC-------ACUCGCCGCCA-AUCGACGAACGCCUAAGCAUCGAAAUUAGCGCAAAUAACAAUUAAAAAUGCUUGAAUGCAUUUAAUUA .(((.(((.((.....((((--....)))).-------....)).))).-.....(((..((....)).)))........)))............((((((......))))))..... ( -23.30) >DroSec_CAF1 18621 108 - 1 AUGCAGGCUAUUUACUGGCG--ACCACGCCC-------ACUAGCCGCCA-AUCGACGAACGCCUAAGCACCGAAAUUAGCGCAAAUAACAAUUAAAAAUGCUUGAAUGCAUUUAAUUA .(((.(((((......((((--....)))).-------..)))))((.(-((...((...((....))..))..))).)))))............((((((......))))))..... ( -23.20) >DroSim_CAF1 17127 108 - 1 AUGCAGGCUGUUUACUGGCG--ACCACGCCC-------ACUCGCCGCCA-AUCGACGAACGCCUAAGCACCAAAAUUAGCGCAAAUAACAAUUAAAAAUGCUUGAAUGCAUUUAAUUA .(((((((........((((--....)))).-------..(((.((...-..)).)))..))))..((..........)))))............((((((......))))))..... ( -23.00) >DroEre_CAF1 15276 118 - 1 AUGCAGGCUAUUUACGGGCGGGACCACGCCCACUGCCCACUGGCCGCCAAAUCGACGAACGCCUAAGCAACGAAAUUAGCGCAAACAACAAUUAAAAAUGCUUGAAUGCAUUUAAUUA .(((((((.......(((((......)))))...(((....)))................))))..((..........)))))............((((((......))))))..... ( -29.60) >DroYak_CAF1 19078 108 - 1 AUGCAGGCUGUUUACCGGCG--ACCACGCCC-------ACUAGCCGCCA-AUCGACGGACGCCUAAGCACCGAAAUUAGCGCAAACAACAAUUAAAAAUGCUUGAAUGCAUUUAAUUA .(((.(((.(((....((((--....)))).-------...))).))).-.....(((..((....)).)))........)))............((((((......))))))..... ( -26.50) >consensus AUGCAGGCUGUUUACUGGCG__ACCACGCCC_______ACUAGCCGCCA_AUCGACGAACGCCUAAGCACCGAAAUUAGCGCAAAUAACAAUUAAAAAUGCUUGAAUGCAUUUAAUUA .(((((((........((((......))))..............((.(.....).))...))))..((..........)))))............((((((......))))))..... (-19.88 = -19.88 + 0.00)

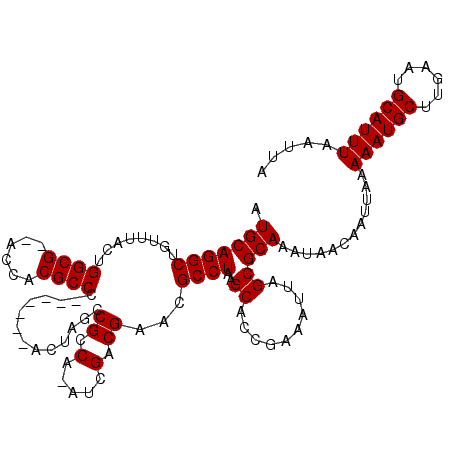
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:31 2006