
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,393,635 – 16,393,798 |
| Length | 163 |
| Max. P | 0.999645 |

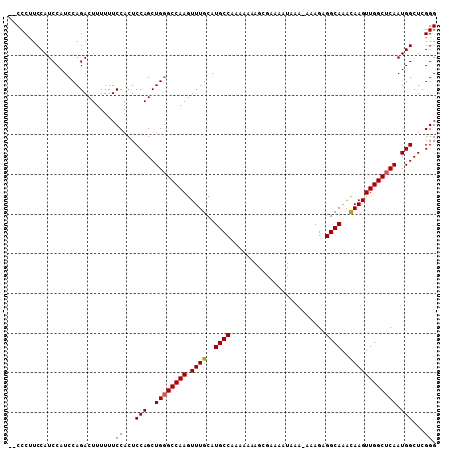
| Location | 16,393,635 – 16,393,729 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 87.54 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -21.68 |
| Energy contribution | -21.79 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.77 |
| Structure conservation index | 0.97 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16393635 94 + 22407834 CCCCCCUCCAUCCAUCCAGACUUUUUUCCACUCCAGCUGUGCCAAGUUUGCAUGCCAAAAA--------------AAAGCGGCAAACAAGUUGGCUCAAUGGCUCGGG ...........................((...(((..((.(((((((((((.(((......--------------...)))))))))...))))).)).)))...)). ( -21.60) >DroSec_CAF1 18183 105 + 1 --CCCUUCCAUCCAUCCAGACUUUUUUCCACUCCAGCUGGGCCAAGUUUGCAUGCCAAAAAAAGCGAAAAUAAA-AAAGAGGCAAAUAAGUUGGCUCAAUGGCUCGGG --.........................((...(((..((((((((.((....((((..................-.....))))...)).)))))))).)))...)). ( -22.20) >DroSim_CAF1 16688 106 + 1 --CCCUUCCAUCCAUCCAGACUUUUUUCCACUCCAGCUGGGCCAAGUUUGCAUGCCAAAAAAAGCGAAAAUAAAAAAAGAGGCAAACAAGUUGGCUCAAUGGCUCGGG --.........................((...(((..((((((((.((((..((((........................))))..)))))))))))).)))...)). ( -23.56) >consensus __CCCUUCCAUCCAUCCAGACUUUUUUCCACUCCAGCUGGGCCAAGUUUGCAUGCCAAAAAAAGCGAAAAUAAA_AAAGAGGCAAACAAGUUGGCUCAAUGGCUCGGG ...........................((...(((..((((((((.((((..((((........................))))..)))))))))))).)))...)). (-21.68 = -21.79 + 0.11)


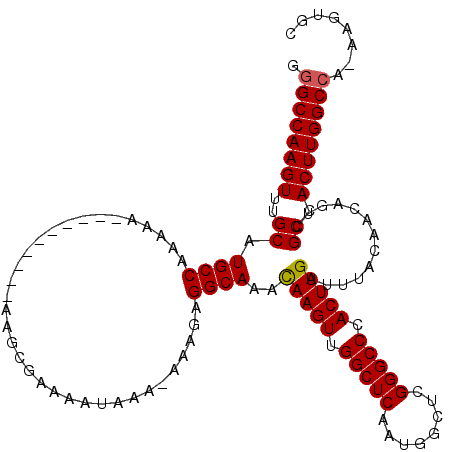
| Location | 16,393,673 – 16,393,768 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.26 |
| Mean single sequence MFE | -33.89 |
| Consensus MFE | -32.26 |
| Energy contribution | -32.30 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.83 |
| SVM RNA-class probability | 0.999645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16393673 95 + 22407834 GUGCCAAGUUUGCAUGCCAAAAA------------------------AAAGCGGCAAACAAGUUGGCUCAAUGGCUCGGGCCCACUUGAUUUACAACAGUCGCCACUUGGCCA-AAGUGC (.(((((((..((.((((.....------------------------.....))))..(((((.(((((........))))).))))).............)).)))))))).-...... ( -31.40) >DroSec_CAF1 18219 108 + 1 GGGCCAAGUUUGCAUGCCAAAAA----------AAGCGAAAAUAAA-AAAGAGGCAAAUAAGUUGGCUCAAUGGCUCGGGCCCACUUGAUUUACAACAGUCGCCACUUGGCCA-AAGUGC .((((((((((((..........----------..)))).......-.....(((....((((.(((((........))))).))))((((......))))))))))))))).-...... ( -33.20) >DroSim_CAF1 16724 109 + 1 GGGCCAAGUUUGCAUGCCAAAAA----------AAGCGAAAAUAAAAAAAGAGGCAAACAAGUUGGCUCAAUGGCUCGGGCCCACUUGAUUUACAACAGUCGCCACUUGGCCA-AAGUGC .((((((((..((.((((.....----------...................))))..(((((.(((((........))))).))))).............)).)))))))).-...... ( -34.26) >DroEre_CAF1 14848 116 + 1 GGGCCAAGUUUGCAUGCCAAAAAAGUGGGGAAAAAGGGAAAAUAAA--AUAAGGCAAGCAAGUUGGCUCAAUGGCUCGGGCCCACUUGAUUUACAACAGUCGCCACUUGGCCA--AGUGC .((((((((.....(.(((......))).)................--....(((....((((.(((((........))))).))))((((......))))))))))))))).--..... ( -33.80) >DroYak_CAF1 18665 107 + 1 GGGCCAAGUUUGCAUGCCAAAAAG-----------GCGAACAUAAA--AUAAGGCAAACAAGUUGGCUCAAUGGCGCGGGCCCACUUGAUUUACAACAGUCGCCACUUGGCCAAAAGUGC .((((((((.....((((.....)-----------)))........--....(((...(((((.(((((........))))).)))))....((....)).)))))))))))........ ( -36.80) >consensus GGGCCAAGUUUGCAUGCCAAAAA__________AAGCGAAAAUAAA_AAAGAGGCAAACAAGUUGGCUCAAUGGCUCGGGCCCACUUGAUUUACAACAGUCGCCACUUGGCCA_AAGUGC .((((((((..((.((((..................................))))..(((((.(((((........))))).))))).............)).))))))))........ (-32.26 = -32.30 + 0.04)


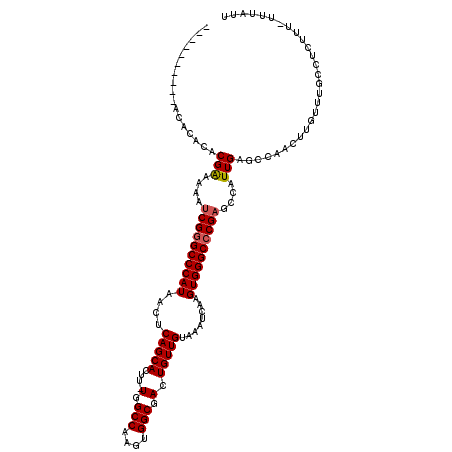
| Location | 16,393,673 – 16,393,768 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.26 |
| Mean single sequence MFE | -34.51 |
| Consensus MFE | -31.00 |
| Energy contribution | -31.24 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16393673 95 - 22407834 GCACUU-UGGCCAAGUGGCGACUGUUGUAAAUCAAGUGGGCCCGAGCCAUUGAGCCAACUUGUUUGCCGCUUU------------------------UUUUUGGCAUGCAAACUUGGCAC ......-..((((((((((.((....))...((((...(((....))).)))))))...((((.(((((....------------------------....))))).))))))))))).. ( -32.40) >DroSec_CAF1 18219 108 - 1 GCACUU-UGGCCAAGUGGCGACUGUUGUAAAUCAAGUGGGCCCGAGCCAUUGAGCCAACUUAUUUGCCUCUUU-UUUAUUUUCGCUU----------UUUUUGGCAUGCAAACUUGGCCC ......-.(((((((((((((..((((....((((...(((....))).))))..))))....))))).....-.........(((.----------.....)))......)))))))). ( -33.40) >DroSim_CAF1 16724 109 - 1 GCACUU-UGGCCAAGUGGCGACUGUUGUAAAUCAAGUGGGCCCGAGCCAUUGAGCCAACUUGUUUGCCUCUUUUUUUAUUUUCGCUU----------UUUUUGGCAUGCAAACUUGGCCC ......-.((((((((.(((...(..(((((.(((((.(((.(((....))).))).))))))))))..).............(((.----------.....))).)))..)))))))). ( -34.80) >DroEre_CAF1 14848 116 - 1 GCACU--UGGCCAAGUGGCGACUGUUGUAAAUCAAGUGGGCCCGAGCCAUUGAGCCAACUUGCUUGCCUUAU--UUUAUUUUCCCUUUUUCCCCACUUUUUUGGCAUGCAAACUUGGCCC .....--.(((((((((((.((....))...((((...(((....))).)))))))...((((.((((....--............................)))).)))))))))))). ( -34.55) >DroYak_CAF1 18665 107 - 1 GCACUUUUGGCCAAGUGGCGACUGUUGUAAAUCAAGUGGGCCCGCGCCAUUGAGCCAACUUGUUUGCCUUAU--UUUAUGUUCGC-----------CUUUUUGGCAUGCAAACUUGGCCC ........((((((((((((((.((((....((((...(((....))).))))..))))..).)))))....--....(((..((-----------(.....)))..))).)))))))). ( -37.40) >consensus GCACUU_UGGCCAAGUGGCGACUGUUGUAAAUCAAGUGGGCCCGAGCCAUUGAGCCAACUUGUUUGCCUCUUU_UUUAUUUUCGCUU__________UUUUUGGCAUGCAAACUUGGCCC ........(((((((((((.((....))...((((...(((....))).)))))))...((((.((((..................................)))).)))))))))))). (-31.00 = -31.24 + 0.24)


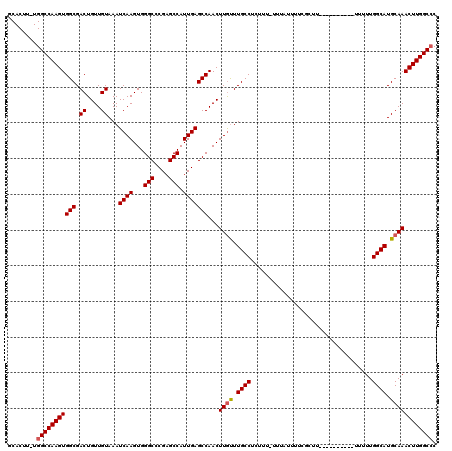
| Location | 16,393,696 – 16,393,798 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.91 |
| Mean single sequence MFE | -30.26 |
| Consensus MFE | -26.94 |
| Energy contribution | -27.18 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.790304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16393696 102 - 22407834 ----------ACACACACGAAAAAUCGGGCCCAUAACUCAGCACUU-UGGCCAAGUGGCGACUGUUGUAAAUCAAGUGGGCCCGAGCCAUUGAGCCAACUUGUUUGCCGCUUU------- ----------...((.((((....((((((((((....(((((...-(.(((....))).).)))))........))))))))))((......))....)))).)).......------- ( -29.90) >DroSec_CAF1 18249 108 - 1 ----------ACACACACGAAAAAUCGGGCCCAUAACUCAGCACUU-UGGCCAAGUGGCGACUGUUGUAAAUCAAGUGGGCCCGAGCCAUUGAGCCAACUUAUUUGCCUCUUU-UUUAUU ----------........((((((((((((((((....(((((...-(.(((....))).).)))))........))))))))))......(((.(((.....))).))))))-)))... ( -31.80) >DroSim_CAF1 16754 109 - 1 ----------ACACACACGAAAAAUCGGGCCCAUAACUCAGCACUU-UGGCCAAGUGGCGACUGUUGUAAAUCAAGUGGGCCCGAGCCAUUGAGCCAACUUGUUUGCCUCUUUUUUUAUU ----------........((((((((((((((((....(((((...-(.(((....))).).)))))........))))))))))......(((.(((.....))).))).))))))... ( -31.90) >DroEre_CAF1 14888 116 - 1 ACACACACACACACACACGGGAAAUCGGGCCCAUAACUCAGCACU--UGGCCAAGUGGCGACUGUUGUAAAUCAAGUGGGCCCGAGCCAUUGAGCCAACUUGCUUGCCUUAU--UUUAUU ...................((...((((((((((....(((((..--(.(((....))).).)))))........)))))))))).))...((((......)))).......--...... ( -34.10) >DroYak_CAF1 18694 108 - 1 ----------ACACACACGAAAAAUCGAGCCCAUAACUCAGCACUUUUGGCCAAGUGGCGACUGUUGUAAAUCAAGUGGGCCCGCGCCAUUGAGCCAACUUGUUUGCCUUAU--UUUAUG ----------...((.((((......(((.......))).......(((((..(((((((.(..((........))..).....)))))))..))))).)))).))......--...... ( -23.60) >consensus __________ACACACACGAAAAAUCGGGCCCAUAACUCAGCACUU_UGGCCAAGUGGCGACUGUUGUAAAUCAAGUGGGCCCGAGCCAUUGAGCCAACUUGUUUGCCUCUUU_UUUAUU .................(((....((((((((((....(((((....(.(((....))).).)))))........))))))))))....)))............................ (-26.94 = -27.18 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:28 2006