
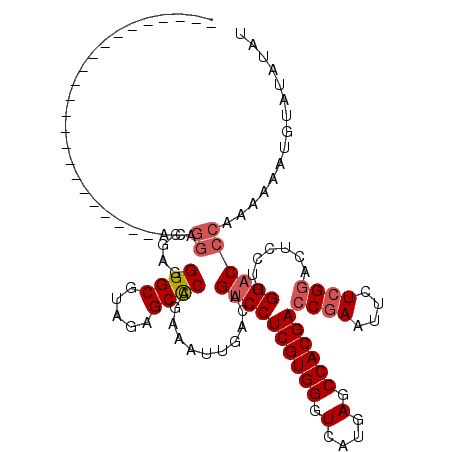
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,392,499 – 16,392,630 |
| Length | 131 |
| Max. P | 0.739895 |

| Location | 16,392,499 – 16,392,596 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.20 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -23.28 |
| Energy contribution | -24.12 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16392499 97 + 22407834 -----------------------ACGGACGAGGGGCGUAGAGCCCAGAAAUUGACAGACCUCGUGGGUCAUGAGCCACGACCGAAUUCUCGGACUCCUUGGACCCAAAAAAAGUAUAUAU -----------------------..(..((((((((.....))))...............((((((.(....).))))))((((....))))....))))..)................. ( -27.10) >DroSec_CAF1 17082 97 + 1 -----------------------ACGGACGAGGGGCGUAGAGCCCAGAAAUUGACAGUCCUCGUGGGUCAUGAGCCACGACCGAAUUCUCGGACUCCUUGGACCCAAAAAAUGUAUAUAU -----------------------..((.(((.((((.....)))).....)))...((((((((((.(....).))))))((((....)))).......))))))............... ( -29.70) >DroSim_CAF1 14095 91 + 1 -----------------------------GAGGGGCGUAGAGCCCAGAAAUUGACAGUCCUCGUGGGUCAUGAGCCACGACCGAAUUCUCGGACUCCUUGGACCCAAAAAAUGUAUAUAU -----------------------------...((((.....))))...........((((((((((.(....).))))))((((....)))).......))))................. ( -27.50) >DroEre_CAF1 13789 120 + 1 CAACGGACAACGGACAACGAACAACGGACGAGGGGCGUAAAGCCCAGAAAUUGACAGUCCUCGUGGGUCAUGAGCCACGACCGAAUUCUGGAACUUCUUGGACCCAAAGAAGGGAAAUAU ...((((...(((............((((...((((.....))))(....).....))))((((((.(....).)))))))))...))))...(((((((....).))))))........ ( -35.00) >DroYak_CAF1 17497 94 + 1 -----------------------AAGGACGAGGGGCGUAAAGCUCAGAAAUUGACAGUCCUCGUGGCUCAUGAGCCACGACCGAAUUCUCGGACUACUCGGACCCAAAAA---GAAAUAU -----------------------..(..((((((((.....))))..........(((((((((((((....))))))))..((....))))))).))))..).......---....... ( -33.30) >consensus _______________________ACGGACGAGGGGCGUAGAGCCCAGAAAUUGACAGUCCUCGUGGGUCAUGAGCCACGACCGAAUUCUCGGACUCCUUGGACCCAAAAAAUGUAUAUAU .........................((.....((((.....))))...........((((((((((.(....).))))))((((....)))).......))))))............... (-23.28 = -24.12 + 0.84)


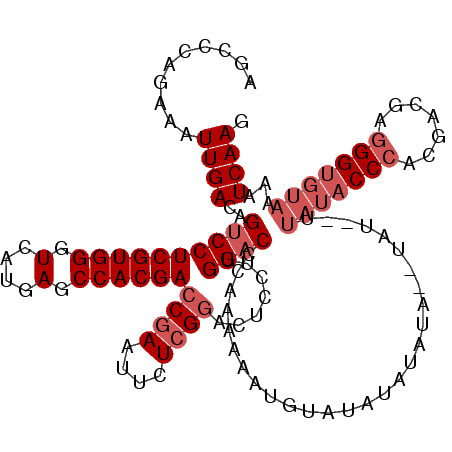
| Location | 16,392,516 – 16,392,630 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.68 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -22.38 |
| Energy contribution | -23.98 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16392516 114 + 22407834 AGCCCAGAAAUUGACAGACCUCGUGGGUCAUGAGCCACGACCGAAUUCUCGGACUCCUUGGACCCAAAAAAAGUAUAUAUA----UAU--UUAUACCCAAGACGAGGGUGUAAAAUCAAG ......((.....(((..(((((((((((..(((.(.(((........)))).)))....))))).......(((((....----...--.))))).....)))))).)))....))... ( -28.60) >DroSec_CAF1 17099 116 + 1 AGCCCAGAAAUUGACAGUCCUCGUGGGUCAUGAGCCACGACCGAAUUCUCGGACUCCUUGGACCCAAAAAAUGUAUAUAUAUA--UAU--UUAUACCCACGACGAGGGUGUAAAAUCAAG ..........((((..((((((((((.(....).))))))((((....)))).......))))....................--..(--((((((((.......))))))))).)))). ( -31.40) >DroSim_CAF1 14106 118 + 1 AGCCCAGAAAUUGACAGUCCUCGUGGGUCAUGAGCCACGACCGAAUUCUCGGACUCCUUGGACCCAAAAAAUGUAUAUAUAUAUGUAU--UUAUACCCACGACGAGGGUGUAAAAUCAAG ..........((((..((((((((((.(....).))))))((((....)))).......))))........................(--((((((((.......))))))))).)))). ( -31.40) >DroEre_CAF1 13829 111 + 1 AGCCCAGAAAUUGACAGUCCUCGUGGGUCAUGAGCCACGACCGAAUUCUGGAACUUCUUGGACCCAAAGAAGGGAAAUAUGUACAC---------CCAGCGGCGAGGGUGUAAAAUCAAG ...((((((.(((.......((((((.(....).)))))).))).)))))).....(((((.(((......))).......(((((---------((........)))))))...))))) ( -34.00) >DroYak_CAF1 17514 117 + 1 AGCUCAGAAAUUGACAGUCCUCGUGGCUCAUGAGCCACGACCGAAUUCUCGGACUACUCGGACCCAAAAA---GAAAUAUGUAUAUACUCGUAUACCCGCGACGAGGGUGUAAAAUCAAG ..........((((..((((((((((((....))))))))((((....)))).......)))).......---..................(((((((.......)))))))...)))). ( -35.00) >consensus AGCCCAGAAAUUGACAGUCCUCGUGGGUCAUGAGCCACGACCGAAUUCUCGGACUCCUUGGACCCAAAAAAUGUAUAUAUAUA__UAU__UUAUACCCACGACGAGGGUGUAAAAUCAAG ..........((((..((((((((((.(....).))))))((((....)))).......))))............................(((((((.......)))))))...)))). (-22.38 = -23.98 + 1.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:23 2006