
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,388,522 – 16,388,681 |
| Length | 159 |
| Max. P | 0.889194 |

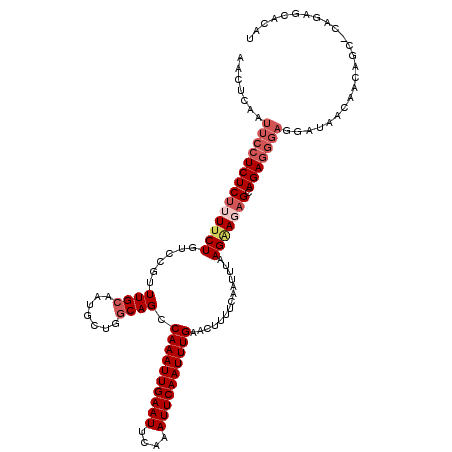
| Location | 16,388,522 – 16,388,641 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.20 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -21.08 |
| Energy contribution | -22.76 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.889194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16388522 119 - 22407834 AACUCAAUUCCUCUCUUUUCUGUCCGUUUGCAACGCUGGCAGCCAAAUUGAAUUCAAAUUCAAUUUGAACUUUUCAAUUUAAGAACAGCAGAGGGAGGCUAACAACAGC-CAGAGCACAU ..(((..(((((((((.((((((((((.....)))..)))...((((((((((....))))))))))..............)))).)).)))))))((((......)))-).)))..... ( -32.60) >DroSec_CAF1 13144 119 - 1 AACUCAAUUCCUCUCUUUUCUGUCCGUUUGCAAUGCUGGCAGCCAAAUUGAAUUCAAAUUCAAUUUGAACUUUUCAAUUUAAGAAGAGCAGAGGGAGGAUAACAACAGC-CAGAGCACAU ........((((((((((.(((((.((.......)).))))).((((((((((....))))))))))..((((((.......)))))).))))))))))........((-....)).... ( -31.70) >DroSim_CAF1 10163 119 - 1 AACUCAAUUCCUCUCUUUUCUGUCCGUUUGCAAUGCUGCCAGCCAAAUUGAAUUCAAAUUCAAUUUGAACUUUUCAAUUUAAGGAGAGCAGAGGGAGGAUAACAACAGC-CAGAGCACAU ........((((((((((.((.((((((.(((....))).)))((((((((((....))))))))))...............))).)).))))))))))........((-....)).... ( -35.20) >DroEre_CAF1 10036 103 - 1 AACUCAAUUCCUCUCUUUUCUGUCCAUUUGCAAUG-UGGCAGCCAAAUUGAAUUCAAAUUCAAUUUGAACUUUUCAAUUUGAGCAGAGCAGA-------------CAGC---CAGCGCAU ..................((((..(...(((....-..)))((((((((((((((((((...)))))))...))))))))).)).)..))))-------------..((---....)).. ( -22.70) >DroYak_CAF1 13441 120 - 1 AACUCAAUUCCUCUCUUUUCUGUCCAUUUGCAAUGCUGGCAGCCAAAUUGAAUUCAAAUUCAAUUUGAACUUUUCAAUUUAAGAACUGCAGAGGGAGGAUAAGAACAGCACAGAGCACAU ...((...((((((((((..(((......)))......((((.((((((((((....))))))))))..(((........)))..))))))))))))))...))...((.....)).... ( -28.60) >consensus AACUCAAUUCCUCUCUUUUCUGUCCGUUUGCAAUGCUGGCAGCCAAAUUGAAUUCAAAUUCAAUUUGAACUUUUCAAUUUAAGAAGAGCAGAGGGAGGAUAACAACAGC_CAGAGCACAU .......((((((((((((((......((((.......)))).((((((((((....))))))))))..............))))))).)))))))........................ (-21.08 = -22.76 + 1.68)

| Location | 16,388,561 – 16,388,681 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -21.84 |
| Energy contribution | -22.04 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16388561 120 + 22407834 AAAUUGAAAAGUUCAAAUUGAAUUUGAAUUCAAUUUGGCUGCCAGCGUUGCAAACGGACAGAAAAGAGAGGAAUUGAGUUAACUGCGUAUGUAAUGGGUAGUUCACAGGAUGCAGGCCAU .........(((.((((((((((....)))))))))))))(((.(((((.....(.....)................((.((((((.(((...))).)))))).))..))))).)))... ( -26.00) >DroSec_CAF1 13183 120 + 1 AAAUUGAAAAGUUCAAAUUGAAUUUGAAUUCAAUUUGGCUGCCAGCAUUGCAAACGGACAGAAAAGAGAGGAAUUGAGUUAACUGCGUAUGUAAUGGGUAGUUCACAGGAUGCAGGCCAU .........(((.((((((((((....)))))))))))))(((.(((((.....(.....)................((.((((((.(((...))).)))))).))..))))).)))... ( -26.40) >DroSim_CAF1 10202 106 + 1 AAAUUGAAAAGUUCAAAUUGAAUUUGAAUUCAAUUUGGCUGGCAGCAUUGCAAACGGACAGAAAAGAGAGGAAUUGAGUUAACUGCGUAUGUAAUGGGUA--------------GGCCAU (((((((...((((((((...)))))))))))))))((((.((..(((((((.(((..(((......((....)).......)))))).))))))).)).--------------)))).. ( -26.22) >DroEre_CAF1 10060 119 + 1 AAAUUGAAAAGUUCAAAUUGAAUUUGAAUUCAAUUUGGCUGCCA-CAUUGCAAAUGGACAGAAAAGAGAGGAAUUGAGUUAACUGCGUGUGUAAUGGGUAGUUCGCAGGAUGCAGGACAU ..........(((((((((((((....)))))))))(((((((.-(((((((.(((..(((......((....)).......)))))).)))))))))))))).((.....)).)))).. ( -28.22) >DroYak_CAF1 13481 120 + 1 AAAUUGAAAAGUUCAAAUUGAAUUUGAAUUCAAUUUGGCUGCCAGCAUUGCAAAUGGACAGAAAAGAGAGGAAUUGAGUUAACUGCGUAUGUAAUGGGUAGUUCACAGGAUGCAGGCCAU .........(((.((((((((((....)))))))))))))(((.(((((.(....).....................((.((((((.(((...))).)))))).))..))))).)))... ( -25.90) >consensus AAAUUGAAAAGUUCAAAUUGAAUUUGAAUUCAAUUUGGCUGCCAGCAUUGCAAACGGACAGAAAAGAGAGGAAUUGAGUUAACUGCGUAUGUAAUGGGUAGUUCACAGGAUGCAGGCCAU .............((((((((((....))))))))))((((((..(((((((.(((..(((......((....)).......)))))).))))))))))))).................. (-21.84 = -22.04 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:13 2006